Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2461 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2461 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 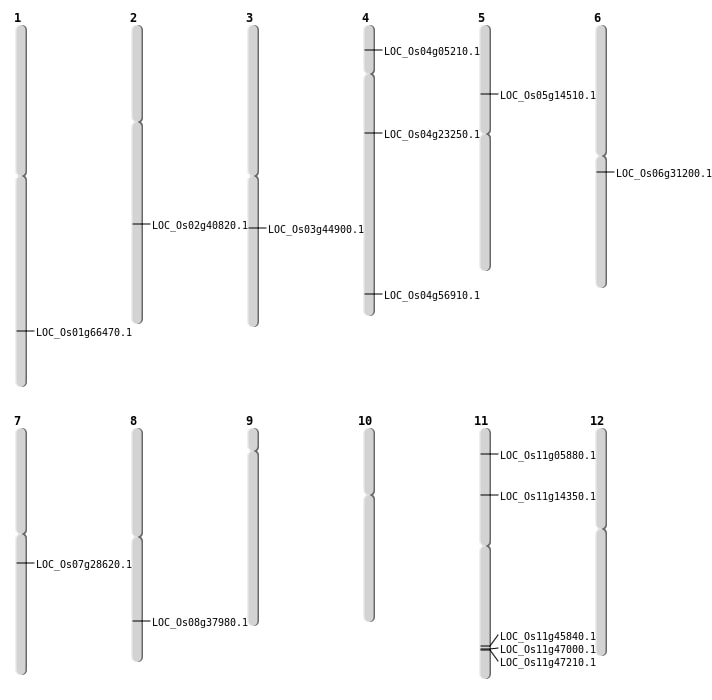
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 25177499 | G-A | HET | ||
| SBS | Chr1 | 26805829 | C-T | HET | ||
| SBS | Chr1 | 28962461 | C-T | HET | ||
| SBS | Chr1 | 30665066 | A-T | HET | ||
| SBS | Chr1 | 38589447 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g66470 |
| SBS | Chr1 | 40306067 | C-T | HET | ||
| SBS | Chr10 | 20830440 | C-A | HET | ||
| SBS | Chr10 | 6151435 | G-A | Homo | ||
| SBS | Chr10 | 9132429 | C-T | Homo | ||
| SBS | Chr11 | 18778658 | G-T | Homo | ||
| SBS | Chr11 | 20276565 | G-T | Homo | ||
| SBS | Chr11 | 20278401 | A-G | Homo | ||
| SBS | Chr11 | 3150644 | C-T | HET | ||
| SBS | Chr11 | 8057968 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g14350 |
| SBS | Chr12 | 15572998 | C-T | Homo | ||
| SBS | Chr12 | 21738862 | C-T | HET | ||
| SBS | Chr12 | 2262725 | C-A | HET | ||
| SBS | Chr12 | 23203341 | T-A | HET | ||
| SBS | Chr12 | 8536028 | G-A | HET | ||
| SBS | Chr2 | 31930692 | C-A | HET | ||
| SBS | Chr2 | 33965025 | T-C | HET | ||
| SBS | Chr2 | 8862708 | G-T | HET | ||
| SBS | Chr3 | 1342082 | T-A | HET | ||
| SBS | Chr3 | 15303993 | C-A | HET | ||
| SBS | Chr3 | 15804765 | T-C | HET | ||
| SBS | Chr3 | 18699629 | G-C | Homo | ||
| SBS | Chr3 | 24144638 | A-G | HET | ||
| SBS | Chr4 | 2311966 | G-A | HET | ||
| SBS | Chr4 | 26669641 | C-A | HET | ||
| SBS | Chr5 | 24800513 | G-A | HET | ||
| SBS | Chr5 | 27263446 | C-T | HET | ||
| SBS | Chr5 | 27526706 | T-C | HET | ||
| SBS | Chr5 | 3099347 | G-A | HET | ||
| SBS | Chr5 | 6390429 | C-T | HET | ||
| SBS | Chr5 | 8210743 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g14510 |
| SBS | Chr6 | 14280865 | T-C | HET | ||
| SBS | Chr6 | 15134547 | T-C | Homo | ||
| SBS | Chr6 | 28019728 | G-A | HET | ||
| SBS | Chr6 | 5406394 | A-T | Homo | ||
| SBS | Chr7 | 12475818 | T-A | Homo | ||
| SBS | Chr7 | 16752200 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g28620 |
| SBS | Chr7 | 22785208 | C-T | HET | ||
| SBS | Chr8 | 12141709 | A-T | HET | ||
| SBS | Chr8 | 14024564 | G-A | HET | ||
| SBS | Chr8 | 18875851 | A-G | HET | ||
| SBS | Chr8 | 24067749 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g37980 |
| SBS | Chr8 | 5735107 | A-T | Homo | ||
| SBS | Chr9 | 12036449 | G-T | HET | ||
| SBS | Chr9 | 19810582 | T-A | HET | ||
| SBS | Chr9 | 5900494 | G-A | HET |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1628741 | 1628743 | 2 | |
| Deletion | Chr12 | 2027626 | 2027627 | 1 | |
| Deletion | Chr4 | 2603080 | 2603085 | 5 | LOC_Os04g05210 |
| Deletion | Chr11 | 2762904 | 2762906 | 2 | LOC_Os11g05880 |
| Deletion | Chr7 | 5301251 | 5301270 | 19 | |
| Deletion | Chr10 | 5832336 | 5832350 | 14 | |
| Deletion | Chr1 | 6259740 | 6259745 | 5 | |
| Deletion | Chr8 | 6590641 | 6590643 | 2 | |
| Deletion | Chr7 | 8300289 | 8300291 | 2 | |
| Deletion | Chr7 | 11251905 | 11251910 | 5 | |
| Deletion | Chr4 | 13265497 | 13265500 | 3 | LOC_Os04g23250 |
| Deletion | Chr8 | 21343971 | 21343977 | 6 | |
| Deletion | Chr2 | 24738784 | 24738794 | 10 | LOC_Os02g40820 |
| Deletion | Chr2 | 33180815 | 33180816 | 1 | |
| Deletion | Chr4 | 33929656 | 33929659 | 3 | LOC_Os04g56910 |
Insertions: 5
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 27732070 | 28401648 | 2 |
| Inversion | Chr11 | 27734982 | 28249119 | 2 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 5168861 | Chr11 | 6458849 | |
| Translocation | Chr6 | 7521744 | Chr3 | 25342952 | 2 |
| Translocation | Chr9 | 8398286 | Chr2 | 25035289 | |
| Translocation | Chr5 | 10026544 | Chr4 | 3220859 | |
| Translocation | Chr9 | 19529882 | Chr6 | 18133104 | 2 |
