Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2572-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2572-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 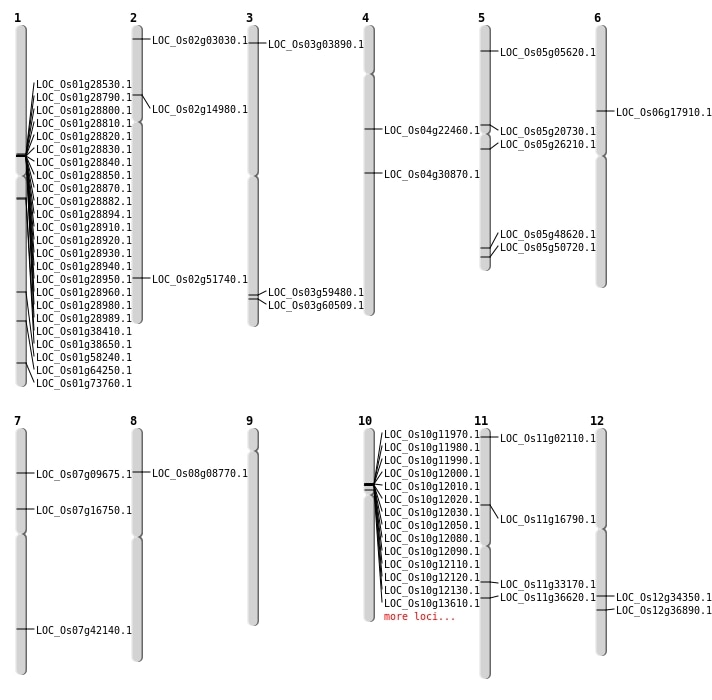
|
| Variant Information |
|---|
Single base substitutions: 114
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10353457 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 11911810 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 13000231 | G-A | HET | ||
| SBS | Chr1 | 15748113 | C-T | Homo | ||
| SBS | Chr1 | 20438343 | C-T | HET | ||
| SBS | Chr1 | 21716978 | T-C | HET | ||
| SBS | Chr1 | 28892527 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 35960102 | A-G | HET | ||
| SBS | Chr1 | 35960102 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 37303883 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g64250 |
| SBS | Chr1 | 39061965 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 42725701 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g73760 |
| SBS | Chr1 | 42761246 | A-G | HET | ||
| SBS | Chr1 | 5995170 | C-T | Homo | ||
| SBS | Chr1 | 5995170 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 620805 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr1 | 676117 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 10738902 | A-T | HET | ||
| SBS | Chr10 | 10738902 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 10739128 | C-T | HET | ||
| SBS | Chr10 | 10739128 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 137396 | G-C | HET | ||
| SBS | Chr10 | 14755863 | A-C | HET | ||
| SBS | Chr10 | 17351029 | C-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 17399066 | G-A | HET | ||
| SBS | Chr10 | 17399066 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 19358309 | A-C | HET | ||
| SBS | Chr10 | 19358309 | A-C | HOMO | SPLICE_SITE_REGION | |
| SBS | Chr10 | 3747862 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 6190529 | G-A | HET | ||
| SBS | Chr10 | 6871509 | C-A | HET | ||
| SBS | Chr10 | 6871509 | C-A | HET | INTRON | |
| SBS | Chr10 | 7379797 | G-A | HET | STOP_GAINED | LOC_Os10g13610 |
| SBS | Chr10 | 7553183 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g13900 |
| SBS | Chr10 | 9844900 | C-G | HET | ||
| SBS | Chr11 | 26451252 | G-A | HET | ||
| SBS | Chr11 | 574224 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g02110 |
| SBS | Chr11 | 9316026 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g16790 |
| SBS | Chr12 | 10592411 | T-G | HET | ||
| SBS | Chr12 | 21866797 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 14223292 | C-A | HET | ||
| SBS | Chr2 | 14223292 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 17756497 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 17994326 | A-G | HET | ||
| SBS | Chr2 | 27004765 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 29461066 | C-T | HET | ||
| SBS | Chr2 | 30937608 | A-G | HET | ||
| SBS | Chr2 | 30937608 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 31964091 | T-G | HET | ||
| SBS | Chr2 | 33114092 | A-T | HET | ||
| SBS | Chr2 | 33114092 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 7756276 | C-T | HET | ||
| SBS | Chr3 | 10144767 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 15816794 | G-A | HET | ||
| SBS | Chr3 | 20448565 | C-A | HET | ||
| SBS | Chr3 | 26935301 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 33854442 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g59480 |
| SBS | Chr3 | 35875684 | G-A | HET | ||
| SBS | Chr3 | 8437723 | A-C | Homo | ||
| SBS | Chr4 | 12714507 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g22460 |
| SBS | Chr4 | 18451218 | G-A | HET | STOP_GAINED | LOC_Os04g30870 |
| SBS | Chr4 | 21186906 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 24221480 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 31631914 | T-C | HET | ||
| SBS | Chr4 | 864734 | C-A | HET | ||
| SBS | Chr5 | 12162741 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g20730 |
| SBS | Chr5 | 14472026 | A-C | HET | ||
| SBS | Chr5 | 17506147 | G-T | HET | ||
| SBS | Chr5 | 17506147 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 18583619 | G-A | HET | ||
| SBS | Chr5 | 18583619 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 24626346 | T-A | HET | ||
| SBS | Chr5 | 24626346 | T-A | HOMO | UTR_3_PRIME | |
| SBS | Chr5 | 24827694 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 26585827 | A-G | HET | ||
| SBS | Chr5 | 27872646 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g48620 |
| SBS | Chr5 | 2797305 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g05620 |
| SBS | Chr5 | 7857729 | T-G | HET | ||
| SBS | Chr5 | 7857732 | T-G | HET | ||
| SBS | Chr5 | 7930264 | T-C | HET | ||
| SBS | Chr6 | 10398457 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g17910 |
| SBS | Chr6 | 10398457 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g17910 |
| SBS | Chr6 | 10604829 | C-T | Homo | ||
| SBS | Chr6 | 10604829 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 17423049 | T-C | Homo | ||
| SBS | Chr6 | 17423049 | T-C | HOMO | INTRON | |
| SBS | Chr6 | 25170058 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 5324569 | C-T | HET | ||
| SBS | Chr7 | 12520786 | C-A | HET | ||
| SBS | Chr7 | 13968602 | C-T | Homo | ||
| SBS | Chr7 | 13968602 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 19779761 | G-A | HET | ||
| SBS | Chr7 | 1981646 | C-G | HET | ||
| SBS | Chr7 | 5291268 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 8902460 | C-G | Homo | ||
| SBS | Chr7 | 8902460 | C-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 9825007 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g16750 |
| SBS | Chr7 | 9825007 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g16750 |
| SBS | Chr8 | 21715424 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5087161 | A-T | HET | STOP_GAINED | LOC_Os08g08770 |
| SBS | Chr8 | 9136073 | G-A | HET | INTRON | |
| SBS | Chr9 | 13351373 | C-T | HET | ||
| SBS | Chr9 | 13351373 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 1926886 | C-A | HET | ||
| SBS | Chr9 | 1926886 | C-A | HOMO | INTRON | |
| SBS | Chr9 | 1926892 | C-A | HET | ||
| SBS | Chr9 | 1926892 | C-A | HOMO | INTRON | |
| SBS | Chr9 | 42844 | C-T | HET | ||
| SBS | Chr9 | 42844 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 4886230 | A-G | HET | ||
| SBS | Chr9 | 4886230 | A-G | HOMO | INTRON | |
| SBS | Chr9 | 4916966 | T-C | HET | ||
| SBS | Chr9 | 6361628 | A-C | HET | ||
| SBS | Chr9 | 91617 | C-T | HET |
Deletions: 41
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 471309 | 471310 | 1 | |
| Deletion | Chr11 | 919717 | 919723 | 6 | |
| Deletion | Chr7 | 930233 | 930242 | 9 | |
| Deletion | Chr1 | 980518 | 980519 | 1 | 2 |
| Deletion | Chr1 | 2043608 | 2043624 | 16 | |
| Deletion | Chr3 | 2582878 | 2582889 | 11 | |
| Deletion | Chr7 | 2906819 | 2906828 | 9 | |
| Deletion | Chr11 | 4470515 | 4470544 | 29 | |
| Deletion | Chr5 | 5636798 | 5636809 | 11 | |
| Deletion | Chr10 | 6634001 | 6748000 | 114000 | 13 |
| Deletion | Chr7 | 6863455 | 6863456 | 1 | |
| Deletion | Chr2 | 8362925 | 8362938 | 13 | 2 |
| Deletion | Chr2 | 9101886 | 9101887 | 1 | |
| Deletion | Chr4 | 9225927 | 9225928 | 1 | 2 |
| Deletion | Chr10 | 10576030 | 10576031 | 1 | 2 |
| Deletion | Chr2 | 10842249 | 10842259 | 10 | |
| Deletion | Chr8 | 10884010 | 10884012 | 2 | |
| Deletion | Chr6 | 11342266 | 11342281 | 15 | 2 |
| Deletion | Chr10 | 11558704 | 11558705 | 1 | |
| Deletion | Chr6 | 13503678 | 13503684 | 6 | 2 |
| Deletion | Chr5 | 13604661 | 13604662 | 1 | 2 |
| Deletion | Chr2 | 14215609 | 14215618 | 9 | 2 |
| Deletion | Chr9 | 14423230 | 14423231 | 1 | |
| Deletion | Chr12 | 15522374 | 15522378 | 4 | 2 |
| Deletion | Chr1 | 16126001 | 16234000 | 108000 | 19 |
| Deletion | Chr2 | 16840196 | 16840204 | 8 | 2 |
| Deletion | Chr9 | 17809574 | 17816333 | 6759 | 2 |
| Deletion | Chr8 | 19820254 | 19820258 | 4 | 2 |
| Deletion | Chr7 | 20781157 | 20781158 | 1 | |
| Deletion | Chr10 | 21562496 | 21562506 | 10 | LOC_Os10g40250 |
| Deletion | Chr12 | 22604673 | 22604677 | 4 | LOC_Os12g36890 |
| Deletion | Chr5 | 28446005 | 28446015 | 10 | |
| Deletion | Chr5 | 28882749 | 28882750 | 1 | |
| Deletion | Chr6 | 29657847 | 29657848 | 1 | |
| Deletion | Chr1 | 30941777 | 30941783 | 6 | |
| Deletion | Chr4 | 31223224 | 31223229 | 5 | 2 |
| Deletion | Chr4 | 31274043 | 31274078 | 35 | 2 |
| Deletion | Chr3 | 35189102 | 35189112 | 10 | |
| Deletion | Chr1 | 35654761 | 35654779 | 18 | 2 |
| Deletion | Chr1 | 36122574 | 36122588 | 14 | 2 |
| Deletion | Chr1 | 38813289 | 38813290 | 1 |
Insertions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 15970530 | 15970530 | 1 | LOC_Os01g28530 |
| Insertion | Chr1 | 39804019 | 39804020 | 2 | |
| Insertion | Chr10 | 817630 | 817630 | 1 | |
| Insertion | Chr12 | 12896716 | 12896716 | 1 | |
| Insertion | Chr12 | 24095494 | 24095495 | 2 | |
| Insertion | Chr2 | 12801310 | 12801310 | 1 | |
| Insertion | Chr2 | 21300284 | 21300284 | 1 | |
| Insertion | Chr3 | 12850614 | 12850614 | 1 | |
| Insertion | Chr5 | 15251210 | 15251210 | 1 | 2 |
| Insertion | Chr5 | 15251210 | 15251210 | 1 | 2 |
| Insertion | Chr5 | 22050941 | 22050941 | 1 | |
| Insertion | Chr5 | 25846306 | 25846307 | 2 | 2 |
| Insertion | Chr5 | 25846306 | 25846307 | 2 | 2 |
| Insertion | Chr5 | 6179376 | 6179377 | 2 | 2 |
| Insertion | Chr5 | 6179376 | 6179377 | 2 | 2 |
| Insertion | Chr5 | 9132863 | 9132863 | 1 | |
| Insertion | Chr7 | 15924046 | 15924046 | 1 | |
| Insertion | Chr7 | 19981738 | 19981738 | 1 | |
| Insertion | Chr8 | 19903555 | 19903555 | 1 |
Inversions: 28
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 1192167 | 1192305 | LOC_Os02g03030 |
| Inversion | Chr3 | 1775160 | 1775665 | LOC_Os03g03890 |
| Inversion | Chr8 | 4584529 | 4584727 | |
| Inversion | Chr7 | 5134366 | 5134578 | LOC_Os07g09675 |
| Inversion | Chr5 | 5363934 | 5364086 | |
| Inversion | Chr11 | 5427562 | 5438438 | |
| Inversion | Chr7 | 5866428 | 5866539 | |
| Inversion | Chr3 | 6343966 | 6344168 | |
| Inversion | Chr10 | 6633601 | 6910090 | |
| Inversion | Chr10 | 6748020 | 6910111 | |
| Inversion | Chr10 | 6748021 | 6910112 | |
| Inversion | Chr3 | 15441861 | 15442058 | |
| Inversion | Chr5 | 18318922 | 18319066 | |
| Inversion | Chr7 | 19348114 | 19348353 | |
| Inversion | Chr11 | 19628838 | 19628951 | LOC_Os11g33170 |
| Inversion | Chr8 | 19895897 | 19896080 | |
| Inversion | Chr12 | 20817617 | 20817820 | LOC_Os12g34350 |
| Inversion | Chr4 | 21065866 | 21066137 | |
| Inversion | Chr1 | 21586694 | 21714906 | 2 |
| Inversion | Chr1 | 21586705 | 21714910 | 2 |
| Inversion | Chr11 | 21611444 | 21611553 | LOC_Os11g36620 |
| Inversion | Chr6 | 23097536 | 23097664 | |
| Inversion | Chr1 | 25070400 | 25070511 | |
| Inversion | Chr7 | 28835958 | 28836181 | |
| Inversion | Chr5 | 29071381 | 29071489 | LOC_Os05g50720 |
| Inversion | Chr4 | 29735556 | 29735785 | |
| Inversion | Chr1 | 33668222 | 33668426 | LOC_Os01g58240 |
| Inversion | Chr1 | 39718336 | 39718675 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 17809574 | Chr7 | 25215217 | 2 |
| Translocation | Chr9 | 17809576 | Chr7 | 25215217 | 2 |
| Translocation | Chr9 | 17816328 | Chr7 | 25215209 | 2 |
| Translocation | Chr12 | 18508134 | Chr2 | 12962842 | |
| Translocation | Chr7 | 21347695 | Chr1 | 16125670 | |
| Translocation | Chr7 | 21349906 | Chr1 | 16234001 | LOC_Os01g28989 |
| Translocation | Chr3 | 34394716 | Chr2 | 31696450 | 2 |
