Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2575-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2575-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 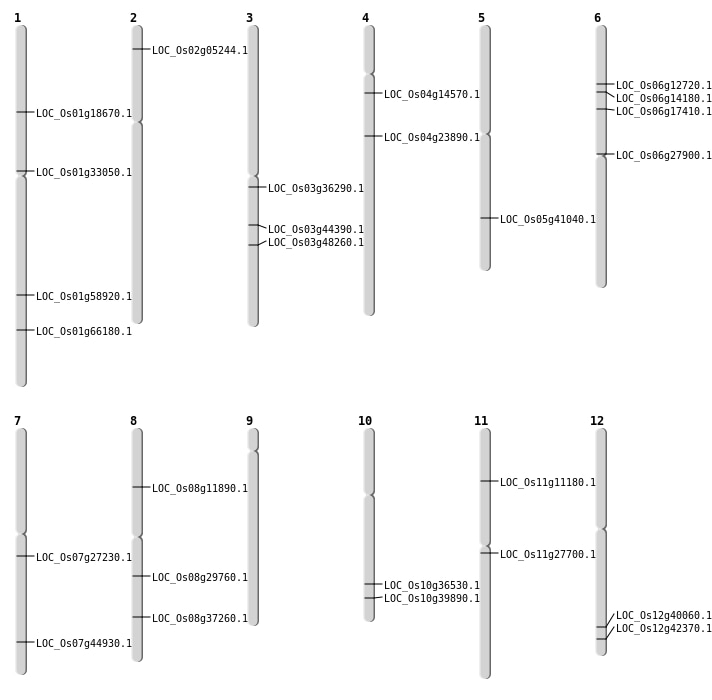
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10520045 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g18670 |
| SBS | Chr1 | 34055245 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g58920 |
| SBS | Chr1 | 34055246 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g58920 |
| SBS | Chr10 | 19546145 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g36530 |
| SBS | Chr10 | 21339204 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g39890 |
| SBS | Chr10 | 4342715 | C-G | Homo | ||
| SBS | Chr11 | 3680537 | A-G | HET | ||
| SBS | Chr12 | 11157391 | C-T | Homo | ||
| SBS | Chr12 | 22593890 | T-C | HET | ||
| SBS | Chr12 | 26336025 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g42370 |
| SBS | Chr12 | 26463238 | T-C | HET | ||
| SBS | Chr2 | 19063024 | T-G | HET | ||
| SBS | Chr3 | 1532732 | C-T | HET | ||
| SBS | Chr3 | 16785655 | C-G | Homo | ||
| SBS | Chr3 | 20128970 | G-A | Homo | STOP_GAINED | LOC_Os03g36290 |
| SBS | Chr3 | 24973245 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g44390 |
| SBS | Chr3 | 25532883 | G-A | Homo | ||
| SBS | Chr3 | 637134 | C-T | HET | ||
| SBS | Chr3 | 9974186 | C-G | Homo | ||
| SBS | Chr4 | 31431238 | C-T | HET | ||
| SBS | Chr4 | 34233484 | G-C | HET | ||
| SBS | Chr4 | 8167326 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g14570 |
| SBS | Chr4 | 9612349 | G-A | HET | ||
| SBS | Chr5 | 13368214 | A-G | HET | ||
| SBS | Chr5 | 28295344 | G-A | HET | ||
| SBS | Chr5 | 29773576 | G-A | HET | ||
| SBS | Chr5 | 5120723 | T-A | HET | ||
| SBS | Chr5 | 9805000 | A-T | HET | ||
| SBS | Chr6 | 17051996 | C-T | Homo | ||
| SBS | Chr6 | 20044181 | G-A | HET | ||
| SBS | Chr6 | 6945200 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g12720 |
| SBS | Chr6 | 7730052 | G-A | HET | ||
| SBS | Chr7 | 17464010 | A-T | HET | ||
| SBS | Chr7 | 29451591 | T-A | HET | ||
| SBS | Chr7 | 630469 | C-A | HET | ||
| SBS | Chr7 | 6476918 | G-A | Homo | ||
| SBS | Chr7 | 6476919 | G-A | Homo | ||
| SBS | Chr8 | 3631743 | T-C | HET | ||
| SBS | Chr8 | 6558540 | T-C | HET | ||
| SBS | Chr8 | 9836022 | G-A | HET | ||
| SBS | Chr9 | 10074488 | C-T | Homo | ||
| SBS | Chr9 | 18472137 | C-T | HET | ||
| SBS | Chr9 | 18472138 | A-T | HET | ||
| SBS | Chr9 | 22528144 | G-C | HET |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 26047 | 26055 | 8 | |
| Deletion | Chr4 | 5802864 | 5802866 | 2 | |
| Deletion | Chr8 | 6968244 | 6968259 | 15 | LOC_Os08g11890 |
| Deletion | Chr3 | 7021194 | 7021200 | 6 | |
| Deletion | Chr3 | 7038059 | 7038061 | 2 | |
| Deletion | Chr6 | 10102297 | 10102309 | 12 | LOC_Os06g17410 |
| Deletion | Chr9 | 11886008 | 11886010 | 2 | |
| Deletion | Chr1 | 17463095 | 17463096 | 1 | |
| Deletion | Chr6 | 19329544 | 19329550 | 6 | |
| Deletion | Chr3 | 19564324 | 19564479 | 155 | |
| Deletion | Chr1 | 19869612 | 19869617 | 5 | |
| Deletion | Chr5 | 20533352 | 20533370 | 18 | |
| Deletion | Chr10 | 23058071 | 23058072 | 1 | |
| Deletion | Chr8 | 23556258 | 23556266 | 8 | LOC_Os08g37260 |
| Deletion | Chr5 | 24051726 | 24051730 | 4 | LOC_Os05g41040 |
| Deletion | Chr12 | 24768586 | 24768587 | 1 | LOC_Os12g40060 |
| Deletion | Chr2 | 30147265 | 30147268 | 3 | |
| Deletion | Chr1 | 41761194 | 41761207 | 13 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 4396816 | 4396817 | 2 |
Inversions: 16
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 2517806 | 2518069 | LOC_Os02g05244 |
| Inversion | Chr4 | 5334643 | 5334922 | |
| Inversion | Chr7 | 7481537 | 7481642 | |
| Inversion | Chr6 | 7916271 | 7916391 | LOC_Os06g14180 |
| Inversion | Chr4 | 8778503 | 8778723 | |
| Inversion | Chr12 | 15568866 | 15568986 | |
| Inversion | Chr7 | 15822493 | 15965744 | LOC_Os07g27230 |
| Inversion | Chr7 | 15822498 | 15965744 | LOC_Os07g27230 |
| Inversion | Chr11 | 15939235 | 15939478 | LOC_Os11g27700 |
| Inversion | Chr1 | 17607763 | 18163258 | 2 |
| Inversion | Chr1 | 17607774 | 18163258 | 2 |
| Inversion | Chr8 | 18294465 | 18294584 | LOC_Os08g29760 |
| Inversion | Chr1 | 22978572 | 22978952 | |
| Inversion | Chr2 | 23530438 | 23530545 | |
| Inversion | Chr5 | 23907016 | 23907317 | |
| Inversion | Chr7 | 26805901 | 26806056 | LOC_Os07g44930 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 6190465 | Chr3 | 27458659 | 2 |
| Translocation | Chr4 | 9441843 | Chr1 | 38446244 | 2 |
| Translocation | Chr4 | 9442257 | Chr1 | 38446247 | 2 |
| Translocation | Chr11 | 14213819 | Chr6 | 15837266 | 2 |
| Translocation | Chr9 | 22697266 | Chr4 | 13654268 | 2 |
| Translocation | Chr9 | 22697287 | Chr4 | 13654191 | 2 |
| Translocation | Chr11 | 27777504 | Chr4 | 9441837 |
