Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2583-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2583-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 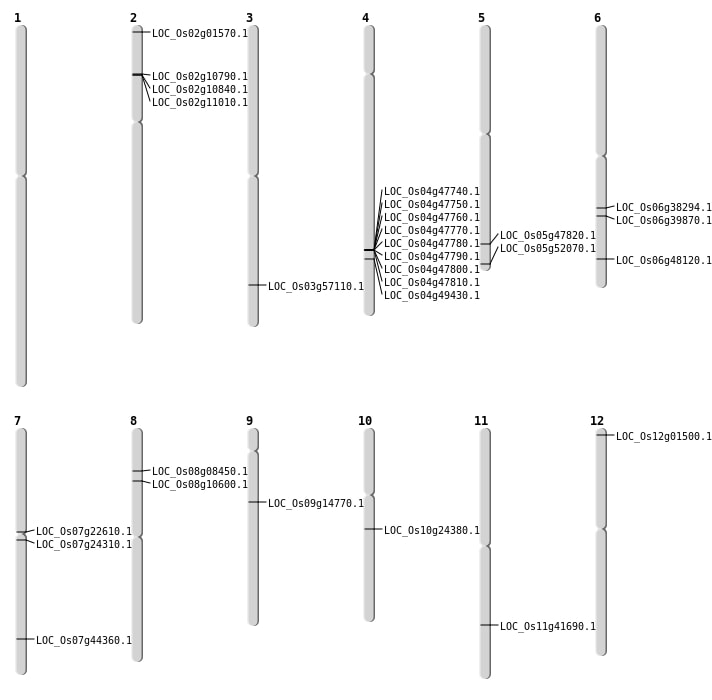
|
| Variant Information |
|---|
Single base substitutions: 76
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17336894 | C-A | HET | ||
| SBS | Chr1 | 24550580 | A-C | HET | ||
| SBS | Chr1 | 27027747 | C-A | HET | ||
| SBS | Chr1 | 3804015 | G-A | HET | ||
| SBS | Chr1 | 4833269 | C-T | HET | ||
| SBS | Chr1 | 6602734 | C-G | HET | ||
| SBS | Chr1 | 9451083 | G-A | HET | ||
| SBS | Chr10 | 12488793 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24380 |
| SBS | Chr10 | 16340448 | G-T | HET | ||
| SBS | Chr10 | 3308970 | G-A | HET | ||
| SBS | Chr10 | 4726640 | T-C | HET | ||
| SBS | Chr10 | 8802673 | C-T | HET | ||
| SBS | Chr11 | 8821862 | C-G | HET | ||
| SBS | Chr12 | 1360469 | A-G | HET | ||
| SBS | Chr12 | 23657294 | C-A | HET | ||
| SBS | Chr12 | 303901 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g01500 |
| SBS | Chr12 | 8662854 | C-T | HET | ||
| SBS | Chr12 | 8662856 | A-G | HET | ||
| SBS | Chr2 | 14766577 | C-T | HET | ||
| SBS | Chr2 | 15198824 | C-G | HET | ||
| SBS | Chr2 | 19409541 | A-G | HET | ||
| SBS | Chr2 | 29434708 | C-T | HET | ||
| SBS | Chr2 | 31197018 | G-T | HET | ||
| SBS | Chr2 | 34215943 | G-A | HET | ||
| SBS | Chr2 | 4963963 | A-T | Homo | ||
| SBS | Chr2 | 5710079 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g10790 |
| SBS | Chr2 | 6951249 | C-T | HET | ||
| SBS | Chr2 | 8952415 | C-T | HET | ||
| SBS | Chr2 | 9388938 | G-A | Homo | ||
| SBS | Chr3 | 17808701 | A-T | HET | ||
| SBS | Chr3 | 19861301 | C-T | HET | ||
| SBS | Chr3 | 27488528 | T-A | HET | ||
| SBS | Chr3 | 27942215 | C-T | HET | ||
| SBS | Chr3 | 32561407 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g57110 |
| SBS | Chr3 | 34427427 | G-T | HET | ||
| SBS | Chr3 | 5797897 | G-A | HET | ||
| SBS | Chr4 | 12566420 | T-A | HET | ||
| SBS | Chr4 | 28151187 | G-A | HET | ||
| SBS | Chr4 | 29498642 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g49430 |
| SBS | Chr4 | 29982430 | G-A | HET | ||
| SBS | Chr4 | 7768785 | T-C | HET | ||
| SBS | Chr4 | 8895581 | G-A | HET | ||
| SBS | Chr5 | 11382103 | C-T | HET | ||
| SBS | Chr5 | 2157761 | G-C | HET | ||
| SBS | Chr5 | 27413929 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g47820 |
| SBS | Chr5 | 29898627 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g52070 |
| SBS | Chr5 | 3955950 | G-T | HET | ||
| SBS | Chr5 | 6714836 | C-T | HET | ||
| SBS | Chr6 | 11065887 | C-T | HET | ||
| SBS | Chr6 | 11643008 | C-A | HET | ||
| SBS | Chr6 | 14582375 | G-T | HET | ||
| SBS | Chr6 | 17376354 | C-T | HET | ||
| SBS | Chr6 | 21924057 | C-T | HET | ||
| SBS | Chr6 | 25094689 | A-G | HET | ||
| SBS | Chr6 | 26150573 | C-A | HET | ||
| SBS | Chr6 | 26150574 | G-A | HET | ||
| SBS | Chr6 | 28726491 | C-T | HET | ||
| SBS | Chr6 | 8735457 | G-T | HET | ||
| SBS | Chr7 | 12727227 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g22610 |
| SBS | Chr7 | 13828211 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g24310 |
| SBS | Chr7 | 20452741 | C-T | HET | ||
| SBS | Chr7 | 25283477 | C-T | HET | ||
| SBS | Chr7 | 7079888 | G-C | HET | ||
| SBS | Chr8 | 16116017 | T-A | HET | ||
| SBS | Chr8 | 19527798 | A-T | HET | ||
| SBS | Chr8 | 22292760 | T-G | HET | ||
| SBS | Chr8 | 4869838 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g08450 |
| SBS | Chr8 | 608259 | T-C | HET | ||
| SBS | Chr8 | 7775581 | A-T | HET | ||
| SBS | Chr8 | 8004312 | C-T | HET | ||
| SBS | Chr9 | 17875778 | A-G | HET | ||
| SBS | Chr9 | 18984900 | G-A | HET | ||
| SBS | Chr9 | 3523158 | G-C | HET | ||
| SBS | Chr9 | 5012363 | C-T | Homo | ||
| SBS | Chr9 | 8097618 | T-C | HET | ||
| SBS | Chr9 | 8787954 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14770 |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 322189 | 322210 | 21 | LOC_Os02g01570 |
| Deletion | Chr3 | 2612107 | 2612117 | 10 | |
| Deletion | Chr7 | 2769658 | 2769660 | 2 | |
| Deletion | Chr8 | 6066930 | 6066941 | 11 | |
| Deletion | Chr8 | 6240437 | 6240448 | 11 | LOC_Os08g10600 |
| Deletion | Chr7 | 6521881 | 6521888 | 7 | |
| Deletion | Chr8 | 9010977 | 9010979 | 2 | |
| Deletion | Chr1 | 9870839 | 9870848 | 9 | |
| Deletion | Chr2 | 10014563 | 10014564 | 1 | |
| Deletion | Chr1 | 10267200 | 10267216 | 16 | |
| Deletion | Chr5 | 12074986 | 12074987 | 1 | |
| Deletion | Chr8 | 13689778 | 13689784 | 6 | |
| Deletion | Chr11 | 17347036 | 17347038 | 2 | |
| Deletion | Chr2 | 19539314 | 19539319 | 5 | |
| Deletion | Chr4 | 21877590 | 21877591 | 1 | |
| Deletion | Chr3 | 22074467 | 22074469 | 2 | |
| Deletion | Chr9 | 22493234 | 22493313 | 79 | |
| Deletion | Chr12 | 22586453 | 22586455 | 2 | |
| Deletion | Chr6 | 23686747 | 23686759 | 12 | LOC_Os06g39870 |
| Deletion | Chr5 | 24728867 | 24728880 | 13 | |
| Deletion | Chr11 | 25030848 | 25030854 | 6 | LOC_Os11g41690 |
| Deletion | Chr4 | 28327001 | 28370000 | 42999 | 8 |
| Deletion | Chr6 | 29116612 | 29116620 | 8 | LOC_Os06g48120 |
| Deletion | Chr6 | 29467616 | 29467622 | 6 | |
| Deletion | Chr2 | 30102415 | 30102416 | 1 | |
| Deletion | Chr4 | 33167103 | 33167106 | 3 | |
| Deletion | Chr2 | 33869258 | 33869276 | 18 |
Insertions: 5
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 5752710 | 5855738 | 2 |
| Inversion | Chr2 | 5752718 | 5855748 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 13766770 | Chr7 | 26506253 | 2 |
| Translocation | Chr6 | 22658312 | Chr1 | 300011 | LOC_Os06g38294 |
