Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2584-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2584-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 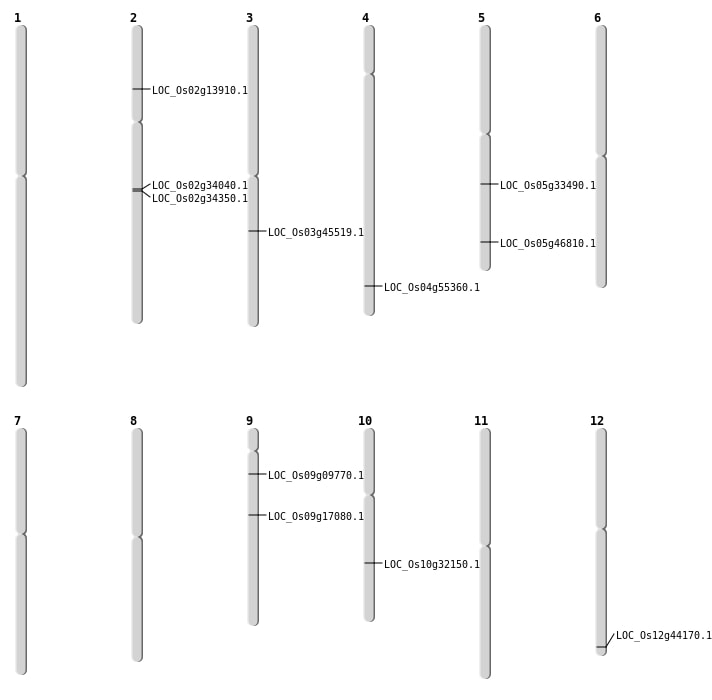
|
| Variant Information |
|---|
Single base substitutions: 47
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18474741 | G-A | Homo | ||
| SBS | Chr1 | 24858136 | G-A | Homo | ||
| SBS | Chr1 | 28643025 | T-A | Homo | ||
| SBS | Chr1 | 32805854 | G-A | Homo | ||
| SBS | Chr1 | 7532638 | C-T | HET | ||
| SBS | Chr10 | 16890741 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g32150 |
| SBS | Chr10 | 18595751 | A-G | HET | ||
| SBS | Chr11 | 10425494 | G-A | HET | ||
| SBS | Chr11 | 25369070 | G-A | Homo | ||
| SBS | Chr11 | 25369071 | C-T | Homo | ||
| SBS | Chr12 | 10006557 | G-T | HET | ||
| SBS | Chr12 | 142298 | C-T | HET | ||
| SBS | Chr12 | 20073900 | G-A | Homo | ||
| SBS | Chr12 | 24410230 | T-A | Homo | ||
| SBS | Chr12 | 27400751 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g44170 |
| SBS | Chr2 | 20341920 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g34040 |
| SBS | Chr2 | 20554307 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g34350 |
| SBS | Chr2 | 24654462 | C-T | HET | ||
| SBS | Chr2 | 30114590 | T-G | HET | ||
| SBS | Chr2 | 32819202 | C-T | Homo | ||
| SBS | Chr2 | 6509402 | G-T | HET | ||
| SBS | Chr2 | 7562007 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g13910 |
| SBS | Chr3 | 18817260 | A-G | Homo | ||
| SBS | Chr3 | 896359 | G-A | HET | ||
| SBS | Chr4 | 15375903 | G-T | HET | ||
| SBS | Chr4 | 19408114 | C-T | Homo | ||
| SBS | Chr4 | 28026251 | A-G | HET | ||
| SBS | Chr4 | 32934422 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g55360 |
| SBS | Chr4 | 5216302 | C-A | HET | ||
| SBS | Chr5 | 10330689 | C-A | HET | ||
| SBS | Chr5 | 19673031 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g33490 |
| SBS | Chr5 | 7981514 | A-C | HET | ||
| SBS | Chr6 | 9691894 | T-A | HET | ||
| SBS | Chr7 | 13397157 | C-A | HET | ||
| SBS | Chr7 | 15750924 | G-T | HET | ||
| SBS | Chr7 | 4188285 | A-G | HET | ||
| SBS | Chr8 | 23102325 | C-T | HET | ||
| SBS | Chr8 | 26372236 | A-G | HET | ||
| SBS | Chr8 | 7695265 | G-A | HET | ||
| SBS | Chr8 | 9753069 | G-A | HET | ||
| SBS | Chr9 | 10463356 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g17080 |
| SBS | Chr9 | 12152297 | G-A | HET | ||
| SBS | Chr9 | 12681397 | A-G | HET | ||
| SBS | Chr9 | 18336102 | G-T | HET | ||
| SBS | Chr9 | 5291882 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g09770 |
| SBS | Chr9 | 6610993 | A-G | HET | ||
| SBS | Chr9 | 8750269 | C-T | HET |
Deletions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 2713369 | 2713370 | 1 | |
| Deletion | Chr9 | 4781481 | 4781482 | 1 | |
| Deletion | Chr5 | 6283066 | 6283069 | 3 | |
| Deletion | Chr3 | 11349347 | 11349355 | 8 | |
| Deletion | Chr1 | 13973794 | 13973813 | 19 | |
| Deletion | Chr5 | 14771817 | 14771831 | 14 | |
| Deletion | Chr11 | 16524575 | 16524583 | 8 | |
| Deletion | Chr2 | 22880317 | 22880327 | 10 | |
| Deletion | Chr5 | 27113934 | 27113945 | 11 | LOC_Os05g46810 |
| Deletion | Chr1 | 27727208 | 27727230 | 22 | |
| Deletion | Chr2 | 34941711 | 34941719 | 8 | |
| Deletion | Chr1 | 39304746 | 39304747 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 20073885 | 20073890 | 6 | |
| Insertion | Chr5 | 29103569 | 29103571 | 3 | |
| Insertion | Chr7 | 8345815 | 8345815 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 781283 | 1757801 | |
| Inversion | Chr10 | 16381339 | 16381970 | |
| Inversion | Chr10 | 16381347 | 16381973 | |
| Inversion | Chr12 | 27416523 | 27416638 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 24390072 | Chr3 | 25692201 | 2 |
