Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2597-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2597-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 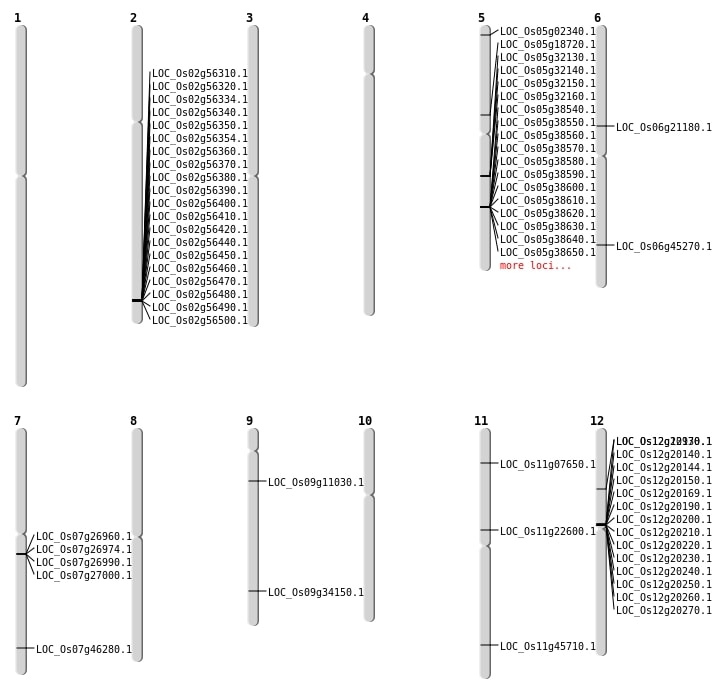
|
| Variant Information |
|---|
Single base substitutions: 63
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17665097 | C-T | HET | ||
| SBS | Chr1 | 17888095 | C-T | HET | ||
| SBS | Chr1 | 23541879 | A-G | HET | ||
| SBS | Chr1 | 25061217 | G-A | HET | ||
| SBS | Chr1 | 34120594 | A-G | HET | ||
| SBS | Chr1 | 41315762 | G-A | HET | ||
| SBS | Chr1 | 9404844 | G-A | HET | ||
| SBS | Chr10 | 17700249 | C-A | HET | ||
| SBS | Chr10 | 19891791 | A-G | Homo | ||
| SBS | Chr10 | 4044289 | G-A | HET | ||
| SBS | Chr10 | 4044290 | G-A | HET | ||
| SBS | Chr10 | 4222896 | G-C | HET | ||
| SBS | Chr10 | 4783811 | A-G | HET | ||
| SBS | Chr10 | 5384461 | G-A | Homo | ||
| SBS | Chr10 | 8129802 | C-T | HET | ||
| SBS | Chr11 | 12998913 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22600 |
| SBS | Chr11 | 28129455 | A-G | HET | ||
| SBS | Chr11 | 28690765 | C-G | HET | ||
| SBS | Chr11 | 3910492 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g07650 |
| SBS | Chr12 | 10816741 | G-A | HET | ||
| SBS | Chr12 | 7187970 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g12970 |
| SBS | Chr12 | 7187971 | G-T | HET | ||
| SBS | Chr2 | 18704727 | T-A | HET | ||
| SBS | Chr2 | 27148022 | A-C | HET | ||
| SBS | Chr2 | 27615608 | T-A | HET | ||
| SBS | Chr2 | 32719836 | T-G | HET | ||
| SBS | Chr3 | 14372758 | C-A | HET | ||
| SBS | Chr3 | 20052203 | G-A | HET | ||
| SBS | Chr3 | 32589428 | G-A | HET | ||
| SBS | Chr3 | 4163767 | T-G | HET | ||
| SBS | Chr3 | 6414708 | G-A | Homo | ||
| SBS | Chr3 | 7253616 | C-A | HET | ||
| SBS | Chr4 | 15496832 | C-T | HET | ||
| SBS | Chr4 | 16327853 | C-G | HET | ||
| SBS | Chr4 | 1955081 | T-C | HET | ||
| SBS | Chr4 | 30336018 | C-T | HET | ||
| SBS | Chr4 | 32705236 | C-T | HET | ||
| SBS | Chr4 | 34594279 | G-A | HET | ||
| SBS | Chr4 | 6328802 | G-A | HET | ||
| SBS | Chr4 | 8961623 | G-A | HET | ||
| SBS | Chr5 | 18780286 | G-A | HET | ||
| SBS | Chr5 | 20913249 | A-C | HET | ||
| SBS | Chr5 | 22290772 | G-A | HET | ||
| SBS | Chr5 | 24403020 | A-G | HET | ||
| SBS | Chr5 | 26678084 | T-A | HET | ||
| SBS | Chr5 | 8567957 | G-A | HET | ||
| SBS | Chr6 | 12242615 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g21180 |
| SBS | Chr6 | 25323260 | C-A | HET | ||
| SBS | Chr6 | 27361518 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g45270 |
| SBS | Chr6 | 29064918 | T-A | HET | ||
| SBS | Chr6 | 6532982 | G-A | Homo | ||
| SBS | Chr7 | 13890751 | C-T | HET | ||
| SBS | Chr7 | 18889974 | A-G | Homo | ||
| SBS | Chr7 | 18889975 | A-T | Homo | ||
| SBS | Chr7 | 21737095 | T-G | HET | ||
| SBS | Chr7 | 27179346 | G-T | HET | ||
| SBS | Chr7 | 9241995 | G-A | HET | ||
| SBS | Chr8 | 14892455 | G-T | HET | ||
| SBS | Chr9 | 11344302 | C-A | HET | ||
| SBS | Chr9 | 13033388 | T-C | HET | ||
| SBS | Chr9 | 20159504 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g34150 |
| SBS | Chr9 | 21264419 | T-C | HET | ||
| SBS | Chr9 | 4307625 | C-A | HET |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 8840801 | 8840806 | 5 | |
| Deletion | Chr3 | 9431096 | 9431097 | 1 | |
| Deletion | Chr12 | 11709001 | 11838000 | 128999 | 14 |
| Deletion | Chr12 | 12807732 | 12807738 | 6 | |
| Deletion | Chr10 | 14975595 | 14975596 | 1 | |
| Deletion | Chr12 | 15436625 | 15436626 | 1 | |
| Deletion | Chr7 | 15615001 | 15629000 | 13999 | 4 |
| Deletion | Chr5 | 18732001 | 18755000 | 22999 | 4 |
| Deletion | Chr10 | 19721612 | 19721618 | 6 | |
| Deletion | Chr9 | 21689475 | 21689491 | 16 | |
| Deletion | Chr12 | 21792566 | 21792572 | 6 | |
| Deletion | Chr5 | 22615001 | 22666000 | 50999 | 13 |
| Deletion | Chr4 | 24345877 | 24345879 | 2 | |
| Deletion | Chr7 | 27615617 | 27615623 | 6 | LOC_Os07g46280 |
| Deletion | Chr11 | 27654217 | 27654232 | 15 | LOC_Os11g45710 |
| Deletion | Chr7 | 28246719 | 28246720 | 1 | |
| Deletion | Chr5 | 28859164 | 28859168 | 4 | |
| Deletion | Chr5 | 29702144 | 29702152 | 8 | |
| Deletion | Chr2 | 34468001 | 34525000 | 56999 | 10 |
| Deletion | Chr2 | 34527001 | 34547000 | 19999 | 4 |
| Deletion | Chr2 | 34549001 | 34608000 | 58999 | 8 |
Insertions: 6
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 11711022 | 11839438 | |
| Inversion | Chr12 | 11837309 | 11839442 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 6127676 | Chr5 | 754978 | 2 |
| Translocation | Chr9 | 8294683 | Chr2 | 16818162 | |
| Translocation | Chr5 | 10844153 | Chr3 | 25789738 | LOC_Os05g18720 |
| Translocation | Chr3 | 14688079 | Chr1 | 3012894 |
