Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2599-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2599-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 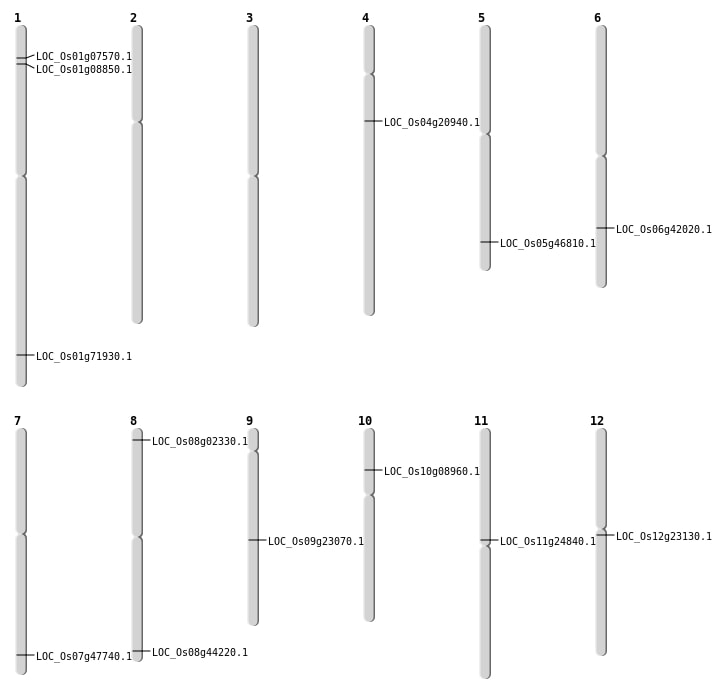
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20210508 | A-G | HET | ||
| SBS | Chr1 | 26702659 | T-C | HET | ||
| SBS | Chr1 | 5807467 | G-A | HET | ||
| SBS | Chr10 | 3149683 | C-T | Homo | ||
| SBS | Chr11 | 14164428 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g24840 |
| SBS | Chr11 | 8512190 | C-G | HET | ||
| SBS | Chr12 | 11321773 | C-T | HET | ||
| SBS | Chr12 | 18716270 | C-G | Homo | ||
| SBS | Chr12 | 19338064 | T-C | HET | ||
| SBS | Chr12 | 25118532 | A-G | HET | ||
| SBS | Chr12 | 5218425 | G-A | HET | ||
| SBS | Chr2 | 14653826 | C-A | HET | ||
| SBS | Chr2 | 24947848 | T-C | HET | ||
| SBS | Chr2 | 2951075 | G-C | HET | ||
| SBS | Chr3 | 23218774 | A-G | Homo | ||
| SBS | Chr3 | 23397017 | C-T | HET | ||
| SBS | Chr3 | 31351250 | C-A | HET | ||
| SBS | Chr3 | 33353143 | A-T | HET | ||
| SBS | Chr3 | 5424274 | A-C | Homo | ||
| SBS | Chr4 | 11751300 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g20940 |
| SBS | Chr4 | 1468273 | T-C | HET | ||
| SBS | Chr4 | 22401918 | C-T | HET | ||
| SBS | Chr4 | 9392478 | C-G | HET | ||
| SBS | Chr5 | 11813285 | G-A | HET | ||
| SBS | Chr5 | 27114453 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g46810 |
| SBS | Chr5 | 29891861 | C-T | HET | ||
| SBS | Chr6 | 25235374 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g42020 |
| SBS | Chr6 | 29445402 | T-C | HET | ||
| SBS | Chr6 | 3292372 | G-A | Homo | ||
| SBS | Chr7 | 28520027 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g47740 |
| SBS | Chr7 | 3999900 | C-T | Homo | ||
| SBS | Chr7 | 8924080 | G-A | HET | ||
| SBS | Chr7 | 9084883 | T-C | Homo | ||
| SBS | Chr7 | 9214351 | G-T | HET | ||
| SBS | Chr8 | 1411741 | C-A | HET | ||
| SBS | Chr9 | 13626653 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g23070 |
| SBS | Chr9 | 4995555 | T-C | HET | ||
| SBS | Chr9 | 5534688 | G-A | HET | ||
| SBS | Chr9 | 8041759 | A-G | HET |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 982852 | 982857 | 5 | |
| Deletion | Chr6 | 1453901 | 1453907 | 6 | |
| Deletion | Chr1 | 4446851 | 4446856 | 5 | LOC_Os01g08850 |
| Deletion | Chr12 | 4769535 | 4769550 | 15 | |
| Deletion | Chr10 | 4847992 | 4848006 | 14 | LOC_Os10g08960 |
| Deletion | Chr3 | 5128076 | 5128077 | 1 | |
| Deletion | Chr7 | 7023960 | 7023962 | 2 | |
| Deletion | Chr5 | 7051244 | 7051257 | 13 | |
| Deletion | Chr3 | 7878580 | 7878583 | 3 | |
| Deletion | Chr10 | 12158976 | 12158978 | 2 | |
| Deletion | Chr12 | 13083986 | 13083987 | 1 | LOC_Os12g23130 |
| Deletion | Chr5 | 13991958 | 13991976 | 18 | |
| Deletion | Chr9 | 19236770 | 19236786 | 16 | |
| Deletion | Chr10 | 21652870 | 21652871 | 1 | |
| Deletion | Chr7 | 28370347 | 28370355 | 8 | |
| Deletion | Chr3 | 33257332 | 33257346 | 14 | |
| Deletion | Chr2 | 35128692 | 35128697 | 5 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 12684453 | 12684462 | 10 | |
| Insertion | Chr6 | 5213412 | 5213427 | 16 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 8077682 | 8367144 | |
| Inversion | Chr3 | 8077685 | 8367149 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 896357 | Chr1 | 41680023 | 2 |
| Translocation | Chr8 | 903055 | Chr1 | 41680034 | 2 |
| Translocation | Chr8 | 10974892 | Chr1 | 3633806 | 2 |
| Translocation | Chr10 | 18634800 | Chr8 | 27836248 | 2 |
