Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2612-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2612-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 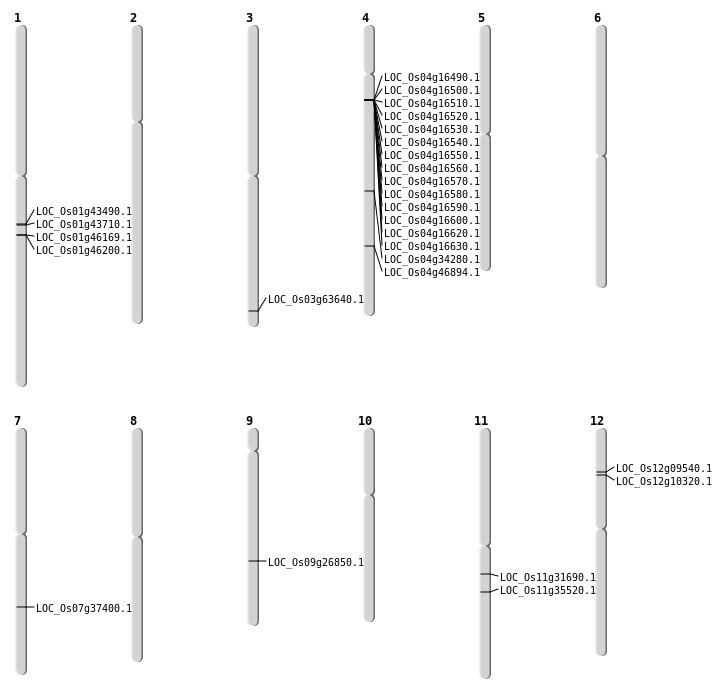
|
| Variant Information |
|---|
Single base substitutions: 61
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14691436 | A-C | HET | ||
| SBS | Chr1 | 18830047 | C-T | HET | ||
| SBS | Chr1 | 21477387 | A-T | HET | ||
| SBS | Chr1 | 27121899 | G-T | HET | ||
| SBS | Chr1 | 27121900 | C-T | HET | ||
| SBS | Chr1 | 34478801 | T-C | Homo | ||
| SBS | Chr1 | 37045416 | G-T | HET | ||
| SBS | Chr1 | 4192688 | C-T | HET | ||
| SBS | Chr10 | 13584422 | A-T | HET | ||
| SBS | Chr10 | 1572530 | T-C | HET | ||
| SBS | Chr10 | 16290743 | C-T | HET | ||
| SBS | Chr10 | 3329305 | C-T | HET | ||
| SBS | Chr10 | 4101760 | G-T | HET | ||
| SBS | Chr10 | 4101761 | T-A | HET | ||
| SBS | Chr10 | 5436806 | T-C | HET | ||
| SBS | Chr11 | 16521096 | G-A | Homo | ||
| SBS | Chr11 | 18660902 | T-A | HET | ||
| SBS | Chr11 | 24304006 | T-A | HET | ||
| SBS | Chr11 | 26404060 | A-G | Homo | ||
| SBS | Chr11 | 26930704 | G-A | HET | ||
| SBS | Chr12 | 10382004 | G-T | Homo | ||
| SBS | Chr12 | 10382005 | C-T | Homo | ||
| SBS | Chr12 | 16343765 | C-T | Homo | ||
| SBS | Chr12 | 27262463 | G-A | HET | ||
| SBS | Chr12 | 27262464 | G-T | HET | ||
| SBS | Chr12 | 5115255 | A-T | HET | ||
| SBS | Chr12 | 5337007 | G-T | HET | ||
| SBS | Chr12 | 8514810 | G-T | HET | ||
| SBS | Chr12 | 9980497 | T-C | Homo | ||
| SBS | Chr2 | 19841183 | G-A | HET | ||
| SBS | Chr2 | 28257157 | A-G | HET | ||
| SBS | Chr3 | 11283919 | A-T | Homo | ||
| SBS | Chr3 | 14633077 | G-A | Homo | ||
| SBS | Chr3 | 32441579 | A-G | HET | ||
| SBS | Chr3 | 35935233 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g63640 |
| SBS | Chr3 | 711061 | G-A | HET | ||
| SBS | Chr4 | 27800164 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g46894 |
| SBS | Chr4 | 31037224 | G-A | HET | ||
| SBS | Chr4 | 69936 | G-A | HET | ||
| SBS | Chr5 | 11399637 | G-A | HET | ||
| SBS | Chr5 | 12845590 | C-A | HET | ||
| SBS | Chr5 | 14438638 | C-A | HET | ||
| SBS | Chr5 | 17477547 | C-T | HET | ||
| SBS | Chr5 | 18642571 | C-A | HET | ||
| SBS | Chr5 | 18912125 | G-A | HET | ||
| SBS | Chr5 | 2245516 | T-G | HET | ||
| SBS | Chr5 | 23186233 | C-T | HET | ||
| SBS | Chr5 | 4803777 | T-C | HET | ||
| SBS | Chr5 | 6338341 | C-T | HET | ||
| SBS | Chr5 | 8810308 | C-A | HET | ||
| SBS | Chr5 | 9064783 | C-T | HET | ||
| SBS | Chr5 | 9296080 | A-G | HET | ||
| SBS | Chr5 | 9664113 | G-A | HET | ||
| SBS | Chr6 | 22546253 | G-A | HET | ||
| SBS | Chr6 | 23833439 | C-T | HET | ||
| SBS | Chr7 | 21347823 | G-A | Homo | ||
| SBS | Chr8 | 14238249 | A-G | HET | ||
| SBS | Chr9 | 11467548 | C-T | HET | ||
| SBS | Chr9 | 14223887 | C-T | HET | ||
| SBS | Chr9 | 16320115 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g26850 |
| SBS | Chr9 | 7090139 | C-T | HET |
Deletions: 21
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 20464452 | 20464459 | 8 | |
| Insertion | Chr5 | 27715782 | 27715789 | 8 | |
| Insertion | Chr7 | 2679762 | 2679763 | 2 | |
| Insertion | Chr8 | 13065401 | 13065401 | 1 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 17802888 | 18236711 | |
| Inversion | Chr11 | 18542219 | 18545371 | 2 |
| Inversion | Chr11 | 20824381 | 20825495 | LOC_Os11g35520 |
| Inversion | Chr1 | 24901355 | 25048239 | 2 |
| Inversion | Chr1 | 25007236 | 25048237 | 2 |
| Inversion | Chr1 | 35182164 | 35667651 | |
| Inversion | Chr1 | 35182166 | 35667652 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 5016429 | Chr4 | 8960522 | LOC_Os12g09540 |
| Translocation | Chr12 | 5456201 | Chr4 | 8960883 | LOC_Os12g10320 |
| Translocation | Chr12 | 6227778 | Chr7 | 22412527 | 2 |
| Translocation | Chr12 | 6510270 | Chr7 | 22412530 | 2 |
| Translocation | Chr7 | 18542793 | Chr2 | 21400253 | |
| Translocation | Chr4 | 20764925 | Chr2 | 20022331 | LOC_Os04g34280 |
| Translocation | Chr4 | 20764934 | Chr2 | 20022188 | LOC_Os04g34280 |
| Translocation | Chr2 | 26568590 | Chr1 | 4970615 |
