Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2637-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2637-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 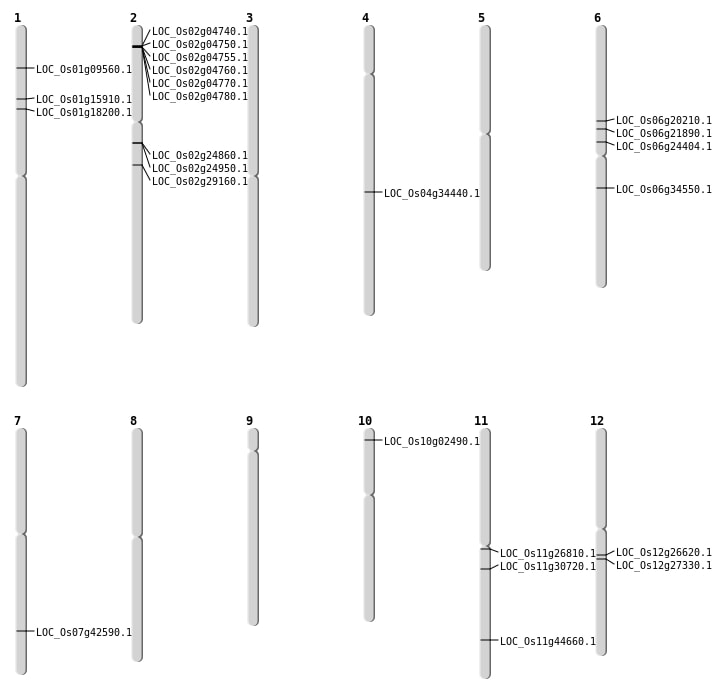
|
| Variant Information |
|---|
Single base substitutions: 58
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16565408 | T-G | Homo | ||
| SBS | Chr1 | 18019721 | G-A | HET | ||
| SBS | Chr1 | 27670077 | T-A | Homo | ||
| SBS | Chr1 | 30842663 | C-G | Homo | ||
| SBS | Chr1 | 3547624 | A-G | HET | ||
| SBS | Chr1 | 37046767 | G-A | HET | ||
| SBS | Chr1 | 43057609 | A-T | HET | ||
| SBS | Chr1 | 8199910 | C-T | HET | ||
| SBS | Chr1 | 9346232 | G-A | HET | ||
| SBS | Chr1 | 9505101 | A-G | HET | ||
| SBS | Chr10 | 21812253 | C-T | HET | ||
| SBS | Chr10 | 22074950 | T-G | Homo | ||
| SBS | Chr10 | 22078664 | C-T | HET | ||
| SBS | Chr10 | 5155337 | T-C | Homo | ||
| SBS | Chr11 | 17891740 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g30720 |
| SBS | Chr11 | 27106521 | C-A | HET | ||
| SBS | Chr11 | 3851324 | T-C | HET | ||
| SBS | Chr11 | 4286116 | T-C | HET | ||
| SBS | Chr11 | 6960635 | C-T | HET | ||
| SBS | Chr12 | 15083607 | G-A | HET | ||
| SBS | Chr12 | 15083608 | C-A | HET | ||
| SBS | Chr12 | 15586441 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g26620 |
| SBS | Chr12 | 16054722 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g27330 |
| SBS | Chr12 | 24272675 | C-T | HET | ||
| SBS | Chr12 | 24592339 | C-T | HET | ||
| SBS | Chr12 | 4740550 | C-T | HET | ||
| SBS | Chr12 | 9129213 | C-T | HET | ||
| SBS | Chr2 | 10880102 | G-T | Homo | ||
| SBS | Chr2 | 14400716 | A-T | HET | ||
| SBS | Chr2 | 14400739 | A-G | HET | ||
| SBS | Chr2 | 14408534 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g24860 |
| SBS | Chr2 | 14467080 | T-A | HET | STOP_GAINED | LOC_Os02g24950 |
| SBS | Chr2 | 17231973 | G-A | HET | ||
| SBS | Chr2 | 2082331 | G-A | HET | ||
| SBS | Chr2 | 2124220 | A-G | HET | ||
| SBS | Chr2 | 22810607 | G-A | Homo | ||
| SBS | Chr2 | 27890958 | G-T | HET | ||
| SBS | Chr2 | 35533033 | G-T | HET | ||
| SBS | Chr2 | 8798496 | T-C | Homo | ||
| SBS | Chr3 | 11154091 | C-A | HET | ||
| SBS | Chr3 | 17178680 | G-A | HET | ||
| SBS | Chr3 | 19407007 | G-A | HET | ||
| SBS | Chr3 | 23342570 | C-T | Homo | ||
| SBS | Chr3 | 23750183 | C-A | Homo | ||
| SBS | Chr3 | 35544934 | T-A | HET | ||
| SBS | Chr4 | 16174321 | A-G | HET | ||
| SBS | Chr4 | 23781948 | C-T | HET | ||
| SBS | Chr4 | 8276191 | G-A | HET | ||
| SBS | Chr5 | 17807760 | G-T | HET | ||
| SBS | Chr5 | 22621524 | C-T | HET | ||
| SBS | Chr5 | 8103257 | C-A | HET | ||
| SBS | Chr5 | 8518069 | T-G | HET | ||
| SBS | Chr7 | 12400244 | G-A | Homo | ||
| SBS | Chr7 | 25489346 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g42590 |
| SBS | Chr8 | 1812306 | G-A | HET | ||
| SBS | Chr8 | 8551709 | G-A | HET | ||
| SBS | Chr9 | 18647927 | C-T | HET | ||
| SBS | Chr9 | 4633442 | A-G | HET |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 2158001 | 2200000 | 41999 | 6 |
| Deletion | Chr4 | 3550049 | 3550061 | 12 | |
| Deletion | Chr10 | 5348997 | 5349007 | 10 | |
| Deletion | Chr5 | 6981773 | 6981775 | 2 | |
| Deletion | Chr6 | 8479240 | 8479241 | 1 | |
| Deletion | Chr12 | 9003502 | 9003522 | 20 | |
| Deletion | Chr10 | 9567287 | 9567288 | 1 | |
| Deletion | Chr6 | 10428913 | 10429013 | 100 | |
| Deletion | Chr1 | 10970963 | 10970965 | 2 | |
| Deletion | Chr2 | 11090905 | 11090914 | 9 | |
| Deletion | Chr6 | 12087683 | 12087684 | 1 | |
| Deletion | Chr10 | 12668053 | 12668061 | 8 | |
| Deletion | Chr2 | 13745225 | 13745236 | 11 | |
| Deletion | Chr9 | 13984592 | 13984599 | 7 | |
| Deletion | Chr1 | 15181772 | 15181775 | 3 | |
| Deletion | Chr1 | 19109652 | 19109653 | 1 | |
| Deletion | Chr12 | 19655723 | 19655731 | 8 | |
| Deletion | Chr6 | 20094842 | 20094844 | 2 | LOC_Os06g34550 |
| Deletion | Chr3 | 23543553 | 23543560 | 7 | |
| Deletion | Chr11 | 27007769 | 27007772 | 3 | LOC_Os11g44660 |
| Deletion | Chr8 | 27457507 | 27457520 | 13 | |
| Deletion | Chr6 | 28766425 | 28766453 | 28 | |
| Deletion | Chr1 | 30842894 | 30842898 | 4 | |
| Deletion | Chr4 | 35280525 | 35280530 | 5 | |
| Deletion | Chr1 | 37671285 | 37671295 | 10 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 32845837 | 32845838 | 2 | |
| Insertion | Chr2 | 9395950 | 9395950 | 1 | |
| Insertion | Chr6 | 8123250 | 8123251 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 4571632 | 4893119 | 2 |
| Inversion | Chr1 | 4571659 | 4893153 | 2 |
| Inversion | Chr1 | 8207829 | 8962770 | 2 |
| Inversion | Chr6 | 11594821 | 12541706 | LOC_Os06g20210 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 924026 | Chr4 | 20857478 | 2 |
| Translocation | Chr10 | 924027 | Chr4 | 20858572 | LOC_Os10g02490 |
| Translocation | Chr8 | 11501495 | Chr1 | 10187287 | 2 |
| Translocation | Chr6 | 12541723 | Chr1 | 8207861 | |
| Translocation | Chr6 | 14285741 | Chr2 | 17279310 | 2 |
| Translocation | Chr6 | 14285763 | Chr2 | 17279598 | 2 |
| Translocation | Chr11 | 15249062 | Chr1 | 11769608 | |
| Translocation | Chr11 | 15372669 | Chr9 | 7355986 | LOC_Os11g26810 |
| Translocation | Chr11 | 15372967 | Chr9 | 7364785 | LOC_Os11g26810 |
| Translocation | Chr12 | 18686678 | Chr5 | 13636098 | |
| Translocation | Chr11 | 22131611 | Chr6 | 12651485 | 2 |
