Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2646-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2646-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 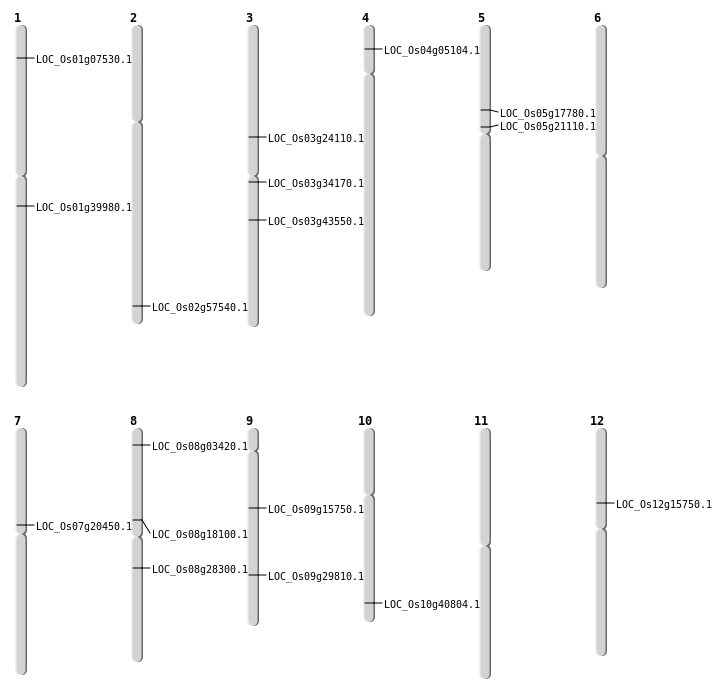
|
| Variant Information |
|---|
Single base substitutions: 58
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20220249 | C-T | HET | ||
| SBS | Chr1 | 2770 | G-T | Homo | ||
| SBS | Chr1 | 34899185 | G-A | HET | ||
| SBS | Chr1 | 38878551 | T-A | Homo | ||
| SBS | Chr1 | 41644748 | G-A | HET | ||
| SBS | Chr1 | 6360742 | T-A | Homo | ||
| SBS | Chr10 | 13888649 | T-A | HET | ||
| SBS | Chr11 | 13236547 | C-A | HET | ||
| SBS | Chr12 | 10582545 | C-T | HET | ||
| SBS | Chr12 | 15877551 | C-T | HET | ||
| SBS | Chr12 | 4620986 | C-T | HET | ||
| SBS | Chr12 | 5494573 | G-A | Homo | ||
| SBS | Chr12 | 8993552 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g15750 |
| SBS | Chr2 | 11214790 | C-T | HET | ||
| SBS | Chr2 | 17599323 | G-T | HET | ||
| SBS | Chr2 | 30848691 | G-T | HET | ||
| SBS | Chr2 | 35257524 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g57540 |
| SBS | Chr2 | 35257528 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g57540 |
| SBS | Chr3 | 13691416 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g24110 |
| SBS | Chr3 | 13759899 | G-C | HET | ||
| SBS | Chr3 | 19458240 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g34170 |
| SBS | Chr3 | 20034556 | G-A | HET | ||
| SBS | Chr3 | 24619645 | T-C | HET | ||
| SBS | Chr3 | 26736385 | A-T | HET | ||
| SBS | Chr3 | 31405544 | C-T | Homo | ||
| SBS | Chr3 | 31446429 | A-G | HET | ||
| SBS | Chr3 | 32336497 | T-C | HET | ||
| SBS | Chr3 | 3955333 | G-A | HET | ||
| SBS | Chr3 | 3955334 | G-A | HET | ||
| SBS | Chr4 | 13105743 | T-G | Homo | ||
| SBS | Chr4 | 13728986 | T-C | Homo | ||
| SBS | Chr4 | 15540237 | C-A | HET | ||
| SBS | Chr4 | 20009650 | G-A | HET | ||
| SBS | Chr5 | 10220132 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17780 |
| SBS | Chr5 | 12413189 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g21110 |
| SBS | Chr5 | 14555606 | G-A | HET | ||
| SBS | Chr5 | 17273591 | G-A | HET | ||
| SBS | Chr5 | 6012010 | C-A | HET | ||
| SBS | Chr5 | 6505219 | C-T | HET | ||
| SBS | Chr6 | 14582280 | C-A | HET | ||
| SBS | Chr6 | 24225913 | C-T | HET | ||
| SBS | Chr7 | 11657218 | C-T | HET | ||
| SBS | Chr7 | 11811435 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g20450 |
| SBS | Chr7 | 15247417 | G-T | HET | ||
| SBS | Chr7 | 2078884 | C-T | HET | ||
| SBS | Chr7 | 2256247 | A-T | HET | ||
| SBS | Chr7 | 2256248 | T-A | HET | ||
| SBS | Chr7 | 2256249 | A-T | HET | ||
| SBS | Chr7 | 25826260 | C-T | HET | ||
| SBS | Chr8 | 1602321 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g03420 |
| SBS | Chr8 | 17272528 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g28300 |
| SBS | Chr8 | 23993810 | C-G | HET | ||
| SBS | Chr8 | 25324816 | A-G | HET | ||
| SBS | Chr8 | 2780851 | C-A | HET | ||
| SBS | Chr8 | 5740774 | C-T | HET | ||
| SBS | Chr8 | 7849030 | C-T | HET | ||
| SBS | Chr9 | 21703402 | C-T | HET | ||
| SBS | Chr9 | 9630881 | C-A | HET |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 3072543 | 3072589 | 46 | |
| Deletion | Chr1 | 3600861 | 3600870 | 9 | LOC_Os01g07530 |
| Deletion | Chr8 | 5337766 | 5337767 | 1 | |
| Deletion | Chr2 | 10597034 | 10597038 | 4 | |
| Deletion | Chr8 | 11096287 | 11096297 | 10 | LOC_Os08g18100 |
| Deletion | Chr4 | 11152010 | 11152021 | 11 | |
| Deletion | Chr2 | 11348649 | 11348653 | 4 | |
| Deletion | Chr6 | 11476300 | 11476301 | 1 | |
| Deletion | Chr7 | 13334459 | 13334473 | 14 | |
| Deletion | Chr9 | 15397337 | 15397338 | 1 | |
| Deletion | Chr9 | 15888272 | 15888278 | 6 | |
| Deletion | Chr9 | 18119528 | 18119532 | 4 | LOC_Os09g29810 |
| Deletion | Chr9 | 21423421 | 21423423 | 2 | |
| Deletion | Chr5 | 24646799 | 24646800 | 1 | |
| Deletion | Chr4 | 25857462 | 25857473 | 11 | |
| Deletion | Chr1 | 34063705 | 34063713 | 8 |
Insertions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 21936603 | 21936603 | 1 | LOC_Os10g40804 |
| Insertion | Chr12 | 3418570 | 3418571 | 2 | |
| Insertion | Chr2 | 20911076 | 20911079 | 4 | |
| Insertion | Chr2 | 35129278 | 35129278 | 1 | |
| Insertion | Chr2 | 6601876 | 6601877 | 2 | |
| Insertion | Chr2 | 8352718 | 8352720 | 3 | |
| Insertion | Chr5 | 5186583 | 5186603 | 21 | |
| Insertion | Chr7 | 17740870 | 17740871 | 2 | |
| Insertion | Chr9 | 18313692 | 18313703 | 12 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 13989774 | 14053245 | |
| Inversion | Chr10 | 13989784 | 14053252 | |
| Inversion | Chr2 | 20632024 | 21342612 | |
| Inversion | Chr2 | 20632032 | 21342627 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 2526788 | Chr3 | 24316453 | 2 |
| Translocation | Chr10 | 3381453 | Chr6 | 25514081 | |
| Translocation | Chr11 | 7492567 | Chr9 | 9620712 | 2 |
| Translocation | Chr11 | 7492575 | Chr9 | 9615444 | |
| Translocation | Chr6 | 25514172 | Chr1 | 22545222 | 2 |
