Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2650-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2650-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 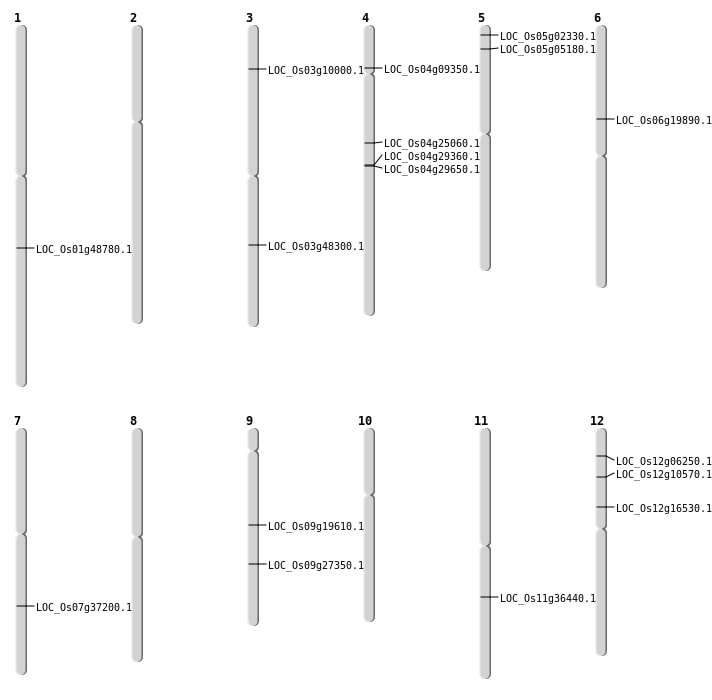
|
| Variant Information |
|---|
Single base substitutions: 62
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16291286 | G-A | HET | ||
| SBS | Chr1 | 23006674 | T-C | HET | ||
| SBS | Chr1 | 27978202 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g48780 |
| SBS | Chr1 | 36199748 | G-C | Homo | ||
| SBS | Chr10 | 20815000 | G-A | Homo | ||
| SBS | Chr10 | 5491435 | C-G | HET | ||
| SBS | Chr11 | 18149131 | G-A | HET | ||
| SBS | Chr11 | 19303668 | T-A | HET | ||
| SBS | Chr11 | 19303670 | G-A | HET | ||
| SBS | Chr11 | 434957 | G-C | Homo | ||
| SBS | Chr11 | 434960 | C-A | Homo | ||
| SBS | Chr12 | 10103382 | C-T | HET | ||
| SBS | Chr12 | 10519292 | G-A | HET | ||
| SBS | Chr12 | 11210205 | A-C | Homo | ||
| SBS | Chr12 | 8308138 | C-T | HET | ||
| SBS | Chr12 | 9474124 | G-A | Homo | STOP_GAINED | LOC_Os12g16530 |
| SBS | Chr2 | 33673084 | G-A | HET | ||
| SBS | Chr2 | 4670248 | T-A | HET | ||
| SBS | Chr3 | 16818251 | T-A | HET | ||
| SBS | Chr3 | 27054328 | T-A | HET | ||
| SBS | Chr3 | 5007270 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g10000 |
| SBS | Chr3 | 9518707 | T-G | Homo | ||
| SBS | Chr4 | 14454723 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g25060 |
| SBS | Chr4 | 16193861 | C-A | HET | ||
| SBS | Chr4 | 1670886 | G-A | HET | ||
| SBS | Chr4 | 17447082 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g29360 |
| SBS | Chr4 | 17649591 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g29650 |
| SBS | Chr4 | 20623312 | G-A | HET | ||
| SBS | Chr4 | 21387349 | G-A | HET | ||
| SBS | Chr4 | 24515635 | A-G | HET | ||
| SBS | Chr4 | 26441922 | A-T | HET | ||
| SBS | Chr4 | 29283325 | G-A | Homo | ||
| SBS | Chr4 | 6595119 | A-G | HET | ||
| SBS | Chr5 | 15023487 | A-G | Homo | ||
| SBS | Chr5 | 15377999 | A-G | Homo | ||
| SBS | Chr5 | 16391825 | G-A | Homo | ||
| SBS | Chr5 | 19851594 | C-T | HET | ||
| SBS | Chr5 | 21157847 | A-C | HET | ||
| SBS | Chr5 | 2355605 | C-A | HET | ||
| SBS | Chr5 | 7054162 | A-C | HET | ||
| SBS | Chr5 | 7206077 | G-A | HET | ||
| SBS | Chr5 | 751149 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g02330 |
| SBS | Chr6 | 10875920 | G-A | HET | ||
| SBS | Chr6 | 11370439 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g19890 |
| SBS | Chr6 | 15215936 | G-C | HET | ||
| SBS | Chr6 | 15460207 | C-G | HET | ||
| SBS | Chr6 | 17776290 | C-T | HET | ||
| SBS | Chr6 | 24698590 | A-T | Homo | ||
| SBS | Chr6 | 27289729 | T-C | HET | ||
| SBS | Chr6 | 8835209 | A-T | HET | ||
| SBS | Chr7 | 10404468 | T-G | HET | ||
| SBS | Chr7 | 10483236 | G-A | HET | ||
| SBS | Chr7 | 18778482 | G-A | HET | ||
| SBS | Chr7 | 18778485 | G-A | HET | ||
| SBS | Chr7 | 22286341 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g37200 |
| SBS | Chr7 | 24219850 | T-C | Homo | ||
| SBS | Chr7 | 8822964 | C-G | HET | ||
| SBS | Chr8 | 25848861 | G-A | HET | ||
| SBS | Chr9 | 11504756 | A-G | HET | ||
| SBS | Chr9 | 13061756 | C-T | HET | ||
| SBS | Chr9 | 16627526 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g27350 |
| SBS | Chr9 | 6985518 | G-T | HET |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 764608 | 764615 | 7 | |
| Deletion | Chr10 | 2508110 | 2508119 | 9 | |
| Deletion | Chr5 | 2524354 | 2524361 | 7 | LOC_Os05g05180 |
| Deletion | Chr11 | 3448563 | 3448565 | 2 | |
| Deletion | Chr12 | 5612246 | 5612251 | 5 | LOC_Os12g10570 |
| Deletion | Chr5 | 7263194 | 7263195 | 1 | |
| Deletion | Chr2 | 9561120 | 9561121 | 1 | |
| Deletion | Chr12 | 12302108 | 12302122 | 14 | |
| Deletion | Chr1 | 14370601 | 14370604 | 3 | |
| Deletion | Chr12 | 14431394 | 14431404 | 10 | |
| Deletion | Chr12 | 14835824 | 14835825 | 1 | |
| Deletion | Chr1 | 15119815 | 15119819 | 4 | |
| Deletion | Chr6 | 21179361 | 21179362 | 1 | |
| Deletion | Chr6 | 23766345 | 23766346 | 1 | |
| Deletion | Chr3 | 27487001 | 27504000 | 16999 | LOC_Os03g48300 |
| Deletion | Chr5 | 28275914 | 28275915 | 1 | |
| Deletion | Chr1 | 32220399 | 32220401 | 2 | |
| Deletion | Chr4 | 34950960 | 34950963 | 3 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23070331 | 23070332 | 2 | |
| Insertion | Chr12 | 4187293 | 4187297 | 5 | |
| Insertion | Chr4 | 11934898 | 11934898 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 3356256 | 3402050 | |
| Inversion | Chr11 | 7223835 | 7665057 | |
| Inversion | Chr11 | 7223853 | 7665076 | |
| Inversion | Chr1 | 9731870 | 9873368 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2973691 | Chr11 | 2085408 | LOC_Os12g06250 |
| Translocation | Chr5 | 11410420 | Chr4 | 4973307 | 2 |
| Translocation | Chr9 | 11727109 | Chr4 | 17810694 | LOC_Os09g19610 |
| Translocation | Chr7 | 12408453 | Chr5 | 3022026 | |
| Translocation | Chr10 | 13367285 | Chr1 | 25286977 | |
| Translocation | Chr11 | 21489132 | Chr1 | 9873365 | LOC_Os11g36440 |
| Translocation | Chr11 | 21489134 | Chr1 | 9872258 | LOC_Os11g36440 |
| Translocation | Chr11 | 26207283 | Chr2 | 30805326 | |
| Translocation | Chr2 | 31842316 | Chr1 | 2441171 |
