Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2653-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2653-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 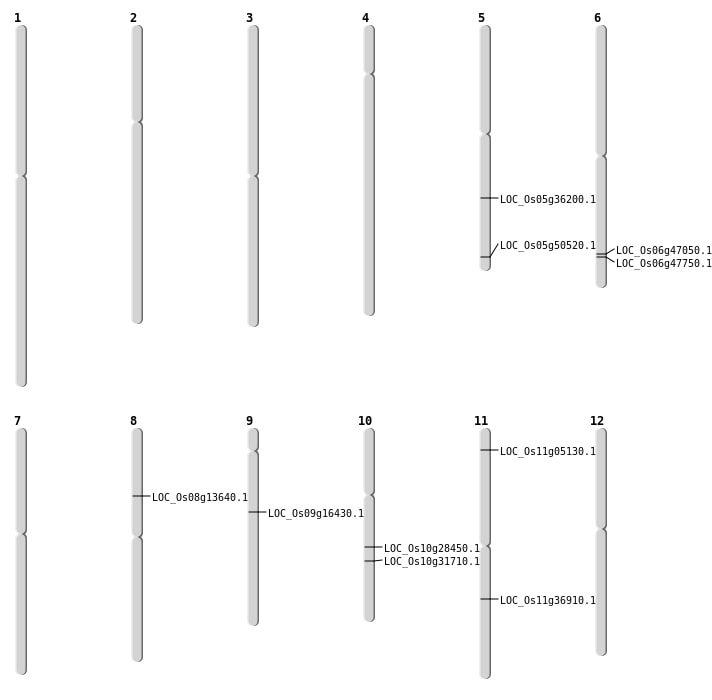
|
| Variant Information |
|---|
Single base substitutions: 51
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19614818 | C-G | HET | ||
| SBS | Chr1 | 23204739 | T-C | HET | ||
| SBS | Chr1 | 26186533 | C-G | Homo | ||
| SBS | Chr1 | 6140844 | G-T | HET | ||
| SBS | Chr10 | 16621183 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g31710 |
| SBS | Chr10 | 18597161 | G-A | Homo | ||
| SBS | Chr10 | 1919697 | T-A | HET | ||
| SBS | Chr10 | 20750109 | A-G | HET | ||
| SBS | Chr11 | 2250610 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g05130 |
| SBS | Chr12 | 10182418 | C-T | HET | ||
| SBS | Chr12 | 15110007 | G-T | HET | ||
| SBS | Chr12 | 16464443 | C-A | HET | ||
| SBS | Chr12 | 4709685 | T-G | HET | ||
| SBS | Chr2 | 17636945 | C-G | HET | ||
| SBS | Chr2 | 17929935 | C-T | Homo | ||
| SBS | Chr2 | 30514806 | G-A | HET | ||
| SBS | Chr2 | 9112646 | A-G | HET | ||
| SBS | Chr3 | 16253217 | T-C | HET | ||
| SBS | Chr3 | 17074992 | C-T | HET | ||
| SBS | Chr3 | 18693802 | A-G | HET | ||
| SBS | Chr3 | 29767303 | A-G | HET | ||
| SBS | Chr3 | 8927398 | C-T | HET | ||
| SBS | Chr3 | 9014695 | G-A | HET | ||
| SBS | Chr4 | 13595416 | C-A | HET | ||
| SBS | Chr4 | 13766709 | G-A | Homo | ||
| SBS | Chr4 | 14093741 | A-C | Homo | ||
| SBS | Chr4 | 2822779 | A-C | HET | ||
| SBS | Chr4 | 6202862 | T-C | HET | ||
| SBS | Chr4 | 8855809 | T-C | HET | ||
| SBS | Chr5 | 13714302 | G-A | HET | ||
| SBS | Chr5 | 14250212 | G-A | HET | ||
| SBS | Chr5 | 16825246 | A-G | HET | ||
| SBS | Chr5 | 21467334 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g36200 |
| SBS | Chr5 | 28975474 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g50520 |
| SBS | Chr5 | 6509435 | G-A | HET | ||
| SBS | Chr6 | 12689291 | C-A | HET | ||
| SBS | Chr6 | 15905220 | A-T | HET | ||
| SBS | Chr6 | 28203501 | C-T | HET | ||
| SBS | Chr6 | 6918524 | G-T | HET | ||
| SBS | Chr7 | 12878602 | C-A | HET | ||
| SBS | Chr7 | 14206830 | G-A | HET | ||
| SBS | Chr7 | 16329976 | G-A | HET | ||
| SBS | Chr7 | 16329977 | T-A | HET | ||
| SBS | Chr7 | 22154035 | G-A | HET | ||
| SBS | Chr8 | 11800725 | G-A | HET | ||
| SBS | Chr8 | 24731424 | T-A | HET | ||
| SBS | Chr8 | 25612749 | C-A | HET | ||
| SBS | Chr8 | 6766423 | C-A | HET | ||
| SBS | Chr8 | 7023837 | C-T | HET | ||
| SBS | Chr9 | 12231184 | A-T | HET | ||
| SBS | Chr9 | 5342695 | C-T | HET |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 885650 | 885651 | 1 | |
| Deletion | Chr3 | 5129481 | 5129482 | 1 | |
| Deletion | Chr6 | 6230951 | 6230959 | 8 | |
| Deletion | Chr3 | 6761968 | 6761977 | 9 | |
| Deletion | Chr9 | 8190293 | 8190299 | 6 | |
| Deletion | Chr10 | 9216474 | 9216476 | 2 | |
| Deletion | Chr1 | 9325105 | 9325109 | 4 | |
| Deletion | Chr2 | 9929738 | 9929750 | 12 | |
| Deletion | Chr9 | 10048792 | 10048793 | 1 | LOC_Os09g16430 |
| Deletion | Chr5 | 12053454 | 12053469 | 15 | |
| Deletion | Chr1 | 12746982 | 12746993 | 11 | |
| Deletion | Chr10 | 14804350 | 14804352 | 2 | LOC_Os10g28450 |
| Deletion | Chr7 | 18697345 | 18697346 | 1 | |
| Deletion | Chr1 | 18731068 | 18731076 | 8 | |
| Deletion | Chr10 | 19134688 | 19134689 | 1 | |
| Deletion | Chr3 | 21217737 | 21217738 | 1 | |
| Deletion | Chr11 | 21768625 | 21768638 | 13 | LOC_Os11g36910 |
| Deletion | Chr1 | 22386899 | 22386901 | 2 | |
| Deletion | Chr1 | 24375933 | 24375935 | 2 | |
| Deletion | Chr4 | 25331268 | 25331284 | 16 | |
| Deletion | Chr8 | 26855194 | 26855213 | 19 | |
| Deletion | Chr1 | 26918799 | 26918801 | 2 | |
| Deletion | Chr6 | 28526001 | 28530000 | 3999 | LOC_Os06g47050 |
| Deletion | Chr3 | 36276133 | 36276136 | 3 |
Insertions: 8
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 7435252 | 8110383 | 2 |
| Inversion | Chr8 | 7435255 | 8110385 | 2 |
| Inversion | Chr6 | 28525860 | 28898525 | 2 |
| Inversion | Chr6 | 28530063 | 28898539 | 2 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 4989751 | Chr9 | 4185128 | |
| Translocation | Chr7 | 18247755 | Chr6 | 28898583 | 2 |
| Translocation | Chr7 | 18247830 | Chr6 | 28530122 | |
| Translocation | Chr11 | 27827382 | Chr5 | 15902467 |
