Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2657-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2657-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 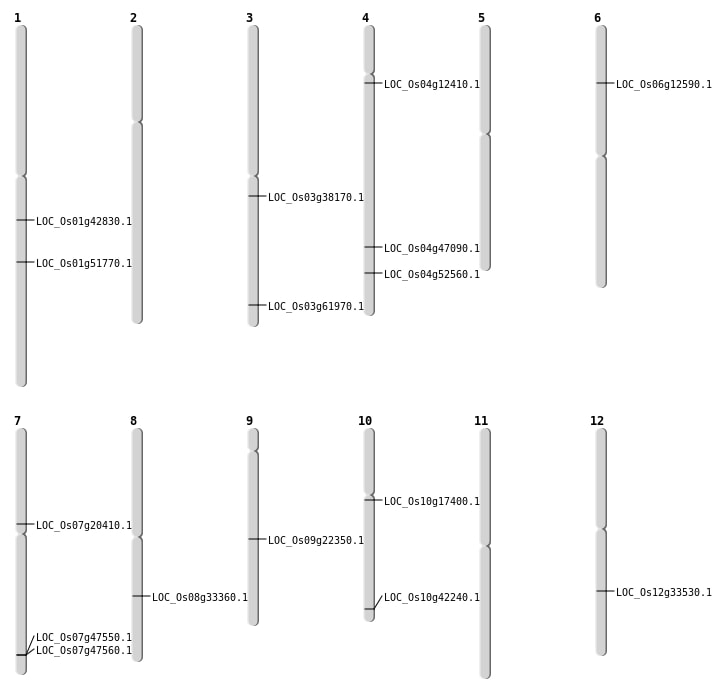
|
| Variant Information |
|---|
Single base substitutions: 76
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12075898 | A-T | HET | ||
| SBS | Chr1 | 18804388 | T-C | HET | ||
| SBS | Chr1 | 24377951 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g42830 |
| SBS | Chr1 | 3131828 | G-T | HET | ||
| SBS | Chr1 | 40423792 | C-T | Homo | ||
| SBS | Chr1 | 440236 | C-T | HET | ||
| SBS | Chr1 | 7031170 | C-T | HET | ||
| SBS | Chr1 | 9560623 | C-G | HET | ||
| SBS | Chr10 | 22411589 | G-A | HET | ||
| SBS | Chr10 | 22751246 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g42240 |
| SBS | Chr10 | 22905633 | G-T | Homo | ||
| SBS | Chr12 | 14045077 | C-T | HET | ||
| SBS | Chr12 | 14928581 | C-A | HET | ||
| SBS | Chr12 | 16117761 | C-T | Homo | ||
| SBS | Chr12 | 16184264 | G-A | HET | ||
| SBS | Chr12 | 20260198 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g33530 |
| SBS | Chr12 | 3759274 | A-G | HET | ||
| SBS | Chr12 | 3995635 | C-T | HET | ||
| SBS | Chr12 | 4407270 | C-A | HET | ||
| SBS | Chr2 | 11652177 | G-A | HET | ||
| SBS | Chr2 | 19517941 | C-T | Homo | ||
| SBS | Chr2 | 34016366 | G-A | HET | ||
| SBS | Chr2 | 35931594 | C-G | Homo | ||
| SBS | Chr3 | 16652106 | G-A | HET | ||
| SBS | Chr3 | 20569314 | C-A | HET | ||
| SBS | Chr3 | 23107164 | A-T | HET | ||
| SBS | Chr3 | 24784694 | A-G | HET | ||
| SBS | Chr3 | 29855228 | G-A | Homo | ||
| SBS | Chr3 | 34773353 | T-A | HET | ||
| SBS | Chr3 | 35127517 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g61970 |
| SBS | Chr3 | 36344507 | T-C | Homo | ||
| SBS | Chr3 | 9517651 | A-T | Homo | ||
| SBS | Chr4 | 12044000 | A-G | HET | ||
| SBS | Chr4 | 13091793 | T-C | Homo | ||
| SBS | Chr4 | 14201602 | T-A | Homo | ||
| SBS | Chr4 | 17895489 | G-T | HET | ||
| SBS | Chr4 | 18940525 | C-T | HET | ||
| SBS | Chr4 | 19462099 | C-T | HET | ||
| SBS | Chr4 | 21810963 | T-C | HET | ||
| SBS | Chr4 | 27961550 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g47090 |
| SBS | Chr4 | 34533603 | A-T | HET | ||
| SBS | Chr4 | 4908894 | C-T | HET | ||
| SBS | Chr4 | 6853547 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g12410 |
| SBS | Chr4 | 9938035 | G-A | HET | ||
| SBS | Chr5 | 17833199 | C-T | HET | ||
| SBS | Chr5 | 5139023 | G-A | HET | ||
| SBS | Chr6 | 11994474 | C-T | HET | ||
| SBS | Chr6 | 14072701 | C-A | HET | ||
| SBS | Chr6 | 30713687 | G-T | HET | ||
| SBS | Chr6 | 30713688 | C-T | HET | ||
| SBS | Chr6 | 30713689 | C-T | HET | ||
| SBS | Chr6 | 5145788 | G-A | HET | ||
| SBS | Chr6 | 6824316 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g12590 |
| SBS | Chr6 | 6824319 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g12590 |
| SBS | Chr6 | 7316597 | C-A | HET | ||
| SBS | Chr6 | 7316598 | T-G | HET | ||
| SBS | Chr6 | 7316599 | A-T | HET | ||
| SBS | Chr6 | 7815983 | G-A | HET | ||
| SBS | Chr6 | 8814749 | T-C | HET | ||
| SBS | Chr7 | 10541787 | T-C | HET | ||
| SBS | Chr7 | 27547363 | C-A | Homo | ||
| SBS | Chr7 | 4283338 | G-A | HET | ||
| SBS | Chr7 | 5554353 | T-C | HET | ||
| SBS | Chr7 | 6183888 | G-T | HET | ||
| SBS | Chr8 | 15471670 | C-G | HET | ||
| SBS | Chr8 | 17242303 | G-A | HET | ||
| SBS | Chr8 | 1806451 | C-T | HET | ||
| SBS | Chr8 | 1896523 | G-T | HET | ||
| SBS | Chr8 | 5057293 | T-A | HET | ||
| SBS | Chr9 | 12265040 | T-A | HET | ||
| SBS | Chr9 | 12672827 | A-G | HET | ||
| SBS | Chr9 | 13507587 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g22350 |
| SBS | Chr9 | 13551576 | C-A | HET | ||
| SBS | Chr9 | 17580736 | T-A | HET | ||
| SBS | Chr9 | 2093671 | T-A | HET | ||
| SBS | Chr9 | 21727308 | C-T | HET |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 4141568 | 4141569 | 1 | |
| Deletion | Chr10 | 5139802 | 5139838 | 36 | |
| Deletion | Chr11 | 5548782 | 5549201 | 419 | |
| Deletion | Chr10 | 7028293 | 7028294 | 1 | |
| Deletion | Chr1 | 8139193 | 8139202 | 9 | |
| Deletion | Chr10 | 8773122 | 8773126 | 4 | LOC_Os10g17400 |
| Deletion | Chr11 | 10280807 | 10280817 | 10 | |
| Deletion | Chr8 | 10446982 | 10446983 | 1 | |
| Deletion | Chr3 | 11237637 | 11237657 | 20 | |
| Deletion | Chr1 | 14258533 | 14258535 | 2 | |
| Deletion | Chr1 | 18819117 | 18819119 | 2 | |
| Deletion | Chr6 | 19498771 | 19498772 | 1 | |
| Deletion | Chr2 | 22735043 | 22735046 | 3 | |
| Deletion | Chr6 | 26584863 | 26584883 | 20 | |
| Deletion | Chr2 | 27358804 | 27358807 | 3 | |
| Deletion | Chr7 | 28438001 | 28445000 | 6999 | 2 |
| Deletion | Chr1 | 29771568 | 29771569 | 1 | LOC_Os01g51770 |
| Deletion | Chr4 | 31250853 | 31250855 | 2 | LOC_Os04g52560 |
| Deletion | Chr3 | 34484716 | 34484721 | 5 | |
| Deletion | Chr2 | 35809575 | 35809580 | 5 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 22105273 | 22105273 | 1 | |
| Insertion | Chr3 | 7417105 | 7417106 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 8106863 | 8685880 | |
| Inversion | Chr11 | 19602278 | 19602386 | |
| Inversion | Chr8 | 20808723 | 20808884 | LOC_Os08g33360 |
| Inversion | Chr3 | 21195069 | 21195353 | LOC_Os03g38170 |
| Inversion | Chr11 | 24908341 | 24908572 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 1330010 | Chr8 | 14822960 | |
| Translocation | Chr11 | 4926344 | Chr7 | 6421522 | |
| Translocation | Chr7 | 11781006 | Chr1 | 20049378 | LOC_Os07g20410 |
| Translocation | Chr7 | 11781174 | Chr1 | 20049379 | LOC_Os07g20410 |
| Translocation | Chr12 | 15976550 | Chr7 | 11781189 | 2 |
| Translocation | Chr12 | 15977128 | Chr7 | 11780997 | 2 |
| Translocation | Chr11 | 25851451 | Chr6 | 16300079 |
