Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2663-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2663-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 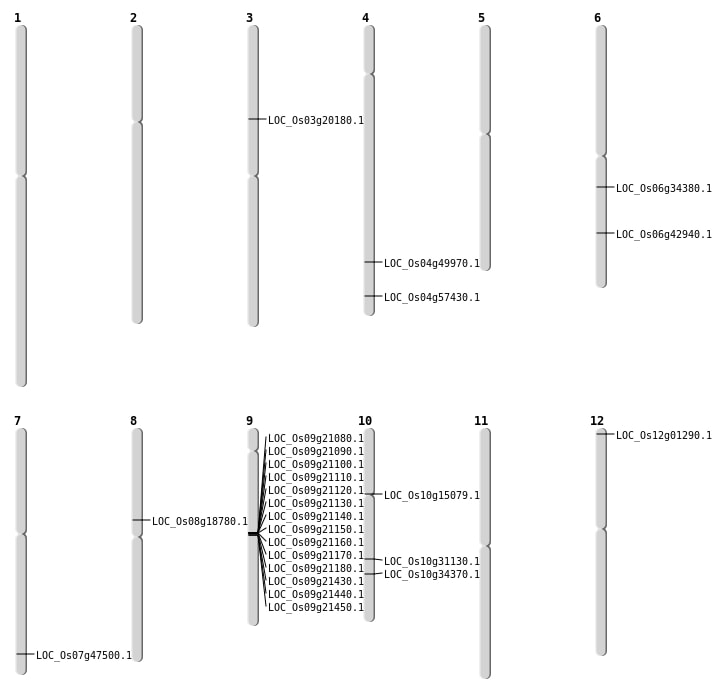
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 40089933 | C-G | HET | ||
| SBS | Chr10 | 16297997 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g31130 |
| SBS | Chr10 | 3924360 | T-C | HET | ||
| SBS | Chr10 | 7899135 | C-T | Homo | ||
| SBS | Chr10 | 7899136 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g15079 |
| SBS | Chr11 | 21009832 | A-C | HET | ||
| SBS | Chr11 | 21548515 | T-A | HET | ||
| SBS | Chr11 | 21700850 | C-T | HET | ||
| SBS | Chr11 | 21700854 | G-T | HET | ||
| SBS | Chr12 | 11760299 | A-T | HET | ||
| SBS | Chr12 | 12425761 | T-A | HET | ||
| SBS | Chr12 | 14298373 | C-T | Homo | ||
| SBS | Chr12 | 168895 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g01290 |
| SBS | Chr12 | 1813115 | C-T | HET | ||
| SBS | Chr12 | 21554003 | A-T | Homo | ||
| SBS | Chr2 | 4353988 | C-T | HET | ||
| SBS | Chr2 | 9767386 | C-T | HET | ||
| SBS | Chr3 | 11406150 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g20180 |
| SBS | Chr3 | 13073337 | G-A | HET | ||
| SBS | Chr3 | 13257933 | G-A | HET | ||
| SBS | Chr3 | 18014668 | G-C | HET | ||
| SBS | Chr3 | 20785466 | C-T | HET | ||
| SBS | Chr3 | 28777327 | G-T | Homo | ||
| SBS | Chr3 | 33041331 | C-T | HET | ||
| SBS | Chr3 | 33589632 | G-T | Homo | ||
| SBS | Chr4 | 16395076 | G-T | HET | ||
| SBS | Chr4 | 17135403 | G-A | HET | ||
| SBS | Chr4 | 17138586 | A-G | HET | ||
| SBS | Chr4 | 29809959 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g49970 |
| SBS | Chr4 | 29809960 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g49970 |
| SBS | Chr4 | 30077013 | A-G | HET | ||
| SBS | Chr4 | 407861 | T-C | Homo | ||
| SBS | Chr5 | 16181265 | G-A | Homo | ||
| SBS | Chr6 | 11197781 | T-A | HET | ||
| SBS | Chr6 | 11197782 | A-T | HET | ||
| SBS | Chr6 | 16565115 | C-T | HET | ||
| SBS | Chr6 | 20008881 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g34380 |
| SBS | Chr6 | 22308469 | C-T | HET | ||
| SBS | Chr6 | 24593113 | C-G | HET | ||
| SBS | Chr6 | 25801440 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g42940 |
| SBS | Chr7 | 16445403 | C-T | Homo | ||
| SBS | Chr8 | 5169462 | G-T | Homo |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 574959 | 574981 | 22 | |
| Deletion | Chr8 | 8368102 | 8368106 | 4 | |
| Deletion | Chr11 | 11136043 | 11136049 | 6 | |
| Deletion | Chr8 | 11209419 | 11210414 | 995 | LOC_Os08g18780 |
| Deletion | Chr3 | 11659702 | 11659703 | 1 | |
| Deletion | Chr9 | 12708001 | 12784000 | 75999 | 11 |
| Deletion | Chr9 | 12956001 | 12975000 | 18999 | 3 |
| Deletion | Chr1 | 14433004 | 14433010 | 6 | |
| Deletion | Chr7 | 15062982 | 15062985 | 3 | |
| Deletion | Chr12 | 15590496 | 15590511 | 15 | |
| Deletion | Chr10 | 16256491 | 16256495 | 4 | |
| Deletion | Chr10 | 17212733 | 17212754 | 21 | |
| Deletion | Chr10 | 18320139 | 18320140 | 1 | LOC_Os10g34370 |
| Deletion | Chr6 | 26837998 | 26838004 | 6 | |
| Deletion | Chr7 | 28401001 | 28407000 | 5999 | LOC_Os07g47500 |
| Deletion | Chr2 | 31215577 | 31215582 | 5 | |
| Deletion | Chr4 | 34171789 | 34171889 | 100 | LOC_Os04g57430 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 29911017 | 29911021 | 5 | |
| Insertion | Chr9 | 939763 | 939763 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 22758949 | 22759643 | |
| Inversion | Chr11 | 22758954 | 22803941 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 8667519 | Chr11 | 23449746 |
