Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2664-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2664-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 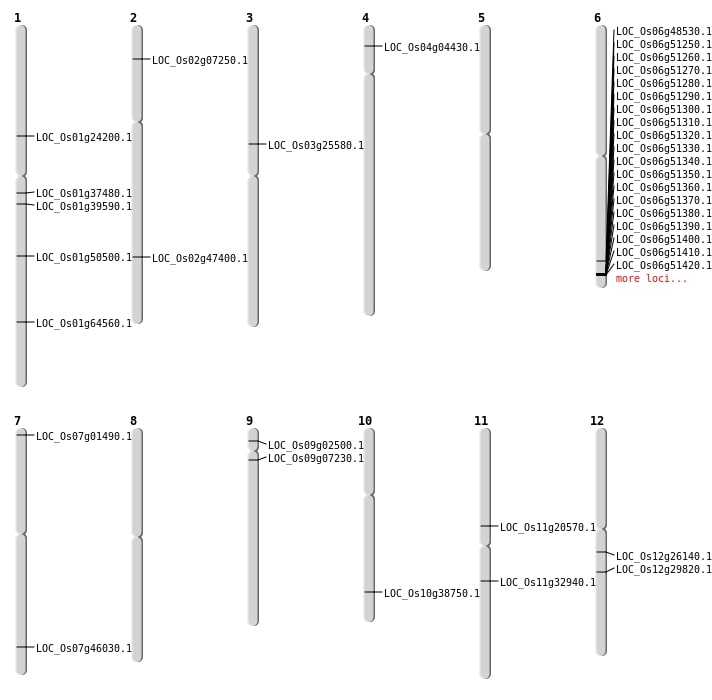
|
| Variant Information |
|---|
Single base substitutions: 57
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10946902 | T-A | HET | ||
| SBS | Chr1 | 16558674 | G-T | Homo | ||
| SBS | Chr1 | 19523791 | T-C | HET | ||
| SBS | Chr1 | 20945656 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g37480 |
| SBS | Chr1 | 25794233 | G-A | HET | ||
| SBS | Chr1 | 29002498 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g50500 |
| SBS | Chr1 | 5126922 | A-G | HET | ||
| SBS | Chr10 | 1417487 | C-T | HET | ||
| SBS | Chr10 | 1417492 | C-G | HET | ||
| SBS | Chr10 | 20642242 | G-A | HET | STOP_GAINED | LOC_Os10g38750 |
| SBS | Chr10 | 21409725 | A-G | HET | ||
| SBS | Chr11 | 11198798 | G-A | HET | ||
| SBS | Chr11 | 11929098 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g20570 |
| SBS | Chr11 | 19451937 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g32940 |
| SBS | Chr11 | 24118545 | A-T | HET | ||
| SBS | Chr12 | 10341670 | G-A | Homo | ||
| SBS | Chr12 | 2913830 | T-C | HET | ||
| SBS | Chr12 | 4703510 | G-A | HET | ||
| SBS | Chr2 | 1870508 | G-T | HET | ||
| SBS | Chr2 | 18855984 | T-C | HET | ||
| SBS | Chr2 | 22994668 | G-A | Homo | ||
| SBS | Chr2 | 29182101 | G-A | HET | ||
| SBS | Chr2 | 528723 | C-G | HET | ||
| SBS | Chr3 | 14638711 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g25580 |
| SBS | Chr3 | 16112905 | T-C | HET | ||
| SBS | Chr3 | 19985517 | A-G | HET | ||
| SBS | Chr3 | 23771096 | A-C | HET | ||
| SBS | Chr3 | 26996863 | G-A | HET | ||
| SBS | Chr3 | 27998939 | G-A | HET | ||
| SBS | Chr3 | 32486708 | T-C | Homo | ||
| SBS | Chr3 | 9690927 | T-A | Homo | ||
| SBS | Chr4 | 22966493 | A-G | HET | ||
| SBS | Chr4 | 28257170 | G-A | HET | ||
| SBS | Chr4 | 34708134 | C-T | HET | ||
| SBS | Chr5 | 11125493 | C-T | HET | ||
| SBS | Chr5 | 13764463 | C-T | HET | ||
| SBS | Chr5 | 17848153 | T-C | HET | ||
| SBS | Chr5 | 24663409 | C-A | Homo | ||
| SBS | Chr5 | 4503488 | G-T | Homo | ||
| SBS | Chr5 | 6096539 | T-C | HET | ||
| SBS | Chr5 | 7022614 | G-A | HET | ||
| SBS | Chr6 | 11648174 | T-A | HET | ||
| SBS | Chr6 | 19306617 | C-T | Homo | ||
| SBS | Chr6 | 2173649 | A-T | Homo | ||
| SBS | Chr6 | 29357890 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g48530 |
| SBS | Chr6 | 8475776 | G-A | HET | ||
| SBS | Chr7 | 11389320 | G-C | HET | ||
| SBS | Chr7 | 15938532 | G-A | HET | ||
| SBS | Chr7 | 25221880 | C-T | HET | ||
| SBS | Chr7 | 4631998 | A-G | HET | ||
| SBS | Chr8 | 10638570 | A-T | HET | ||
| SBS | Chr8 | 7874269 | A-T | HET | ||
| SBS | Chr8 | 8305 | G-A | HET | ||
| SBS | Chr9 | 1036762 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g02500 |
| SBS | Chr9 | 18994308 | G-A | HET | ||
| SBS | Chr9 | 3542439 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07230 |
| SBS | Chr9 | 4907812 | G-T | HET |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 309086 | 309100 | 14 | LOC_Os07g01490 |
| Deletion | Chr9 | 1046435 | 1046443 | 8 | |
| Deletion | Chr4 | 2110938 | 2110958 | 20 | LOC_Os04g04430 |
| Deletion | Chr2 | 3724430 | 3724431 | 1 | LOC_Os02g07250 |
| Deletion | Chr1 | 9811276 | 9811283 | 7 | |
| Deletion | Chr6 | 10548381 | 10548384 | 3 | |
| Deletion | Chr6 | 10629535 | 10629542 | 7 | |
| Deletion | Chr1 | 13635805 | 13635807 | 2 | LOC_Os01g24200 |
| Deletion | Chr1 | 13650857 | 13650858 | 1 | |
| Deletion | Chr1 | 13917032 | 13917035 | 3 | |
| Deletion | Chr12 | 17823338 | 17823341 | 3 | LOC_Os12g29820 |
| Deletion | Chr11 | 18738128 | 18738134 | 6 | |
| Deletion | Chr1 | 22333165 | 22333166 | 1 | LOC_Os01g39590 |
| Deletion | Chr1 | 26136447 | 26136449 | 2 | |
| Deletion | Chr6 | 26876823 | 26876830 | 7 | |
| Deletion | Chr7 | 27470081 | 27470087 | 6 | LOC_Os07g46030 |
| Deletion | Chr11 | 27675576 | 27675590 | 14 | |
| Deletion | Chr6 | 31021001 | 31249000 | 227999 | 34 |
| Deletion | Chr4 | 31760731 | 31760733 | 2 | |
| Deletion | Chr1 | 34020771 | 34020781 | 10 | |
| Deletion | Chr1 | 37460570 | 37460573 | 3 | LOC_Os01g64560 |
| Deletion | Chr1 | 39426188 | 39426189 | 1 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 7045053 | 7045053 | 1 | |
| Insertion | Chr12 | 15228442 | 15228455 | 14 | LOC_Os12g26140 |
| Insertion | Chr12 | 18133661 | 18133661 | 1 | |
| Insertion | Chr12 | 24099542 | 24099545 | 4 | |
| Insertion | Chr3 | 16078768 | 16078769 | 2 | |
| Insertion | Chr4 | 28716399 | 28716400 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 26948038 | 27196377 | |
| Inversion | Chr2 | 28939256 | 29057685 | LOC_Os02g47400 |
| Inversion | Chr2 | 28939257 | 29057692 | LOC_Os02g47400 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 491068 | Chr6 | 31023814 |
