Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2670-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2670-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 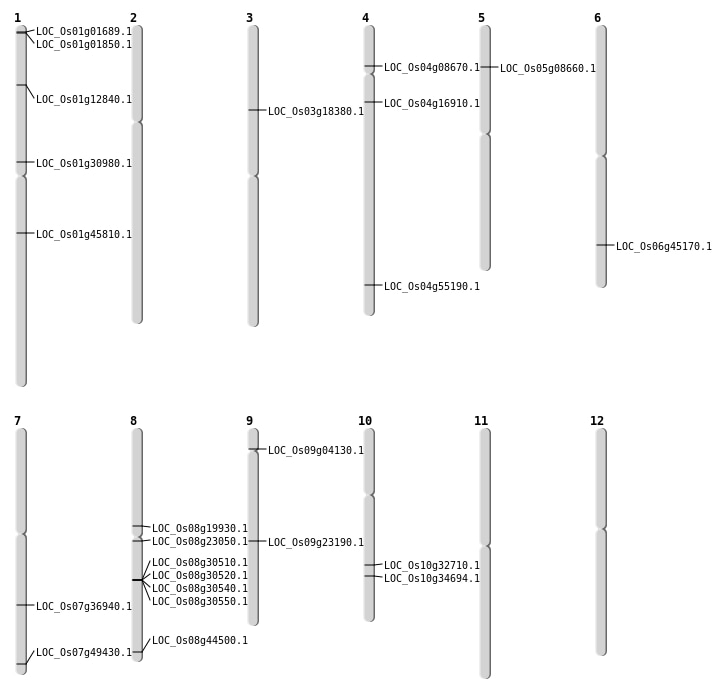
|
| Variant Information |
|---|
Single base substitutions: 71
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10905318 | G-A | HET | ||
| SBS | Chr1 | 13863354 | C-T | HET | ||
| SBS | Chr1 | 16933074 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g30980 |
| SBS | Chr1 | 17516641 | G-A | HET | ||
| SBS | Chr1 | 21565654 | G-T | HET | ||
| SBS | Chr1 | 22310387 | G-T | HET | ||
| SBS | Chr1 | 26026675 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g45810 |
| SBS | Chr1 | 26741041 | A-T | Homo | ||
| SBS | Chr1 | 467591 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g01850 |
| SBS | Chr1 | 5256891 | G-A | HET | ||
| SBS | Chr1 | 7035115 | T-A | HET | ||
| SBS | Chr1 | 7207882 | G-A | HET | ||
| SBS | Chr10 | 18499606 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g34694 |
| SBS | Chr10 | 2048969 | G-A | HET | ||
| SBS | Chr10 | 23184866 | C-T | HET | ||
| SBS | Chr11 | 1302654 | C-T | HET | ||
| SBS | Chr11 | 15306168 | G-A | HET | ||
| SBS | Chr11 | 21616516 | C-T | HET | ||
| SBS | Chr12 | 17334798 | G-A | HET | ||
| SBS | Chr12 | 1997742 | G-T | HET | ||
| SBS | Chr2 | 10449090 | T-C | Homo | ||
| SBS | Chr2 | 10891842 | C-A | Homo | ||
| SBS | Chr2 | 17739096 | A-T | Homo | ||
| SBS | Chr2 | 33536377 | T-C | HET | ||
| SBS | Chr3 | 16122771 | C-T | Homo | ||
| SBS | Chr3 | 16858353 | G-A | HET | ||
| SBS | Chr3 | 18373226 | C-T | Homo | ||
| SBS | Chr3 | 20374514 | A-T | Homo | ||
| SBS | Chr3 | 21547458 | A-C | HET | ||
| SBS | Chr3 | 23945828 | C-T | HET | ||
| SBS | Chr3 | 27670765 | A-T | HET | ||
| SBS | Chr3 | 36191175 | C-T | Homo | ||
| SBS | Chr3 | 4802555 | A-T | HET | ||
| SBS | Chr3 | 902280 | A-G | HET | ||
| SBS | Chr4 | 11966509 | G-A | HET | ||
| SBS | Chr4 | 27631976 | G-T | HET | ||
| SBS | Chr4 | 30349549 | G-T | HET | ||
| SBS | Chr4 | 32809454 | G-T | HET | STOP_GAINED | LOC_Os04g55190 |
| SBS | Chr4 | 9249634 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g16910 |
| SBS | Chr4 | 9861351 | C-T | HET | ||
| SBS | Chr5 | 10345454 | A-T | HET | ||
| SBS | Chr5 | 11719563 | G-A | HET | ||
| SBS | Chr5 | 12886355 | A-C | HET | ||
| SBS | Chr5 | 14870455 | G-A | HET | ||
| SBS | Chr5 | 22083490 | G-T | HET | ||
| SBS | Chr5 | 29045586 | G-A | HET | ||
| SBS | Chr5 | 6581825 | T-C | HET | ||
| SBS | Chr5 | 9860920 | G-A | HET | ||
| SBS | Chr6 | 20183597 | A-T | HET | ||
| SBS | Chr6 | 21931516 | G-T | HET | ||
| SBS | Chr6 | 24595452 | G-A | HET | ||
| SBS | Chr6 | 27312656 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g45170 |
| SBS | Chr6 | 29380851 | C-A | HET | ||
| SBS | Chr6 | 8519236 | C-G | HET | ||
| SBS | Chr7 | 11557878 | A-T | HET | ||
| SBS | Chr7 | 12935761 | C-T | HET | ||
| SBS | Chr7 | 1599876 | T-C | HET | ||
| SBS | Chr7 | 22115446 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36940 |
| SBS | Chr7 | 27582914 | C-T | HET | ||
| SBS | Chr7 | 29601603 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g49430 |
| SBS | Chr8 | 11936344 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g19930 |
| SBS | Chr8 | 2065133 | T-A | HET | ||
| SBS | Chr8 | 27984029 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g44500 |
| SBS | Chr8 | 8531540 | C-T | HET | ||
| SBS | Chr9 | 10101622 | G-A | HET | ||
| SBS | Chr9 | 10425848 | C-T | HET | ||
| SBS | Chr9 | 2145635 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g04130 |
| SBS | Chr9 | 6348303 | G-T | HET | ||
| SBS | Chr9 | 7804645 | C-T | HET | ||
| SBS | Chr9 | 8047827 | C-T | HET | ||
| SBS | Chr9 | 8314823 | C-T | HET |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 69835 | 69836 | 1 | |
| Deletion | Chr1 | 356859 | 356861 | 2 | LOC_Os01g01689 |
| Deletion | Chr6 | 1150471 | 1150480 | 9 | |
| Deletion | Chr8 | 1381500 | 1381501 | 1 | |
| Deletion | Chr1 | 1677066 | 1677075 | 9 | |
| Deletion | Chr8 | 3421359 | 3421360 | 1 | |
| Deletion | Chr1 | 3423610 | 3423615 | 5 | |
| Deletion | Chr5 | 3998081 | 3998090 | 9 | |
| Deletion | Chr4 | 4710361 | 4710372 | 11 | LOC_Os04g08670 |
| Deletion | Chr1 | 7107586 | 7107587 | 1 | LOC_Os01g12840 |
| Deletion | Chr4 | 8073609 | 8073615 | 6 | |
| Deletion | Chr10 | 8365403 | 8365407 | 4 | |
| Deletion | Chr8 | 11416088 | 11416093 | 5 | |
| Deletion | Chr6 | 12275806 | 12275812 | 6 | |
| Deletion | Chr7 | 12960193 | 12960194 | 1 | |
| Deletion | Chr10 | 13126831 | 13126833 | 2 | |
| Deletion | Chr9 | 13749192 | 13749193 | 1 | LOC_Os09g23190 |
| Deletion | Chr5 | 13814225 | 13814238 | 13 | |
| Deletion | Chr8 | 13876905 | 13876934 | 29 | LOC_Os08g23050 |
| Deletion | Chr6 | 14286901 | 14286909 | 8 | |
| Deletion | Chr2 | 17256557 | 17256565 | 8 | |
| Deletion | Chr8 | 18770001 | 18805000 | 34999 | 4 |
| Deletion | Chr3 | 21609473 | 21609475 | 2 | |
| Deletion | Chr1 | 22415418 | 22415422 | 4 | |
| Deletion | Chr8 | 24349849 | 24349850 | 1 | |
| Deletion | Chr1 | 30067058 | 30067059 | 1 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 29949475 | 29949476 | 2 | |
| Insertion | Chr5 | 4740328 | 4740357 | 30 | LOC_Os05g08660 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 2462457 | 2675328 | |
| Inversion | Chr8 | 2462466 | 2676159 | |
| Inversion | Chr3 | 10300795 | 10301112 | LOC_Os03g18380 |
| Inversion | Chr10 | 16478869 | 17124054 | 2 |
| Inversion | Chr10 | 16479154 | 17124055 | 2 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2675333 | Chr2 | 3777780 | |
| Translocation | Chr8 | 9803838 | Chr6 | 9802277 | |
| Translocation | Chr11 | 10341713 | Chr10 | 16478869 | |
| Translocation | Chr11 | 10341715 | Chr10 | 16479149 |
