Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2691-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2691-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 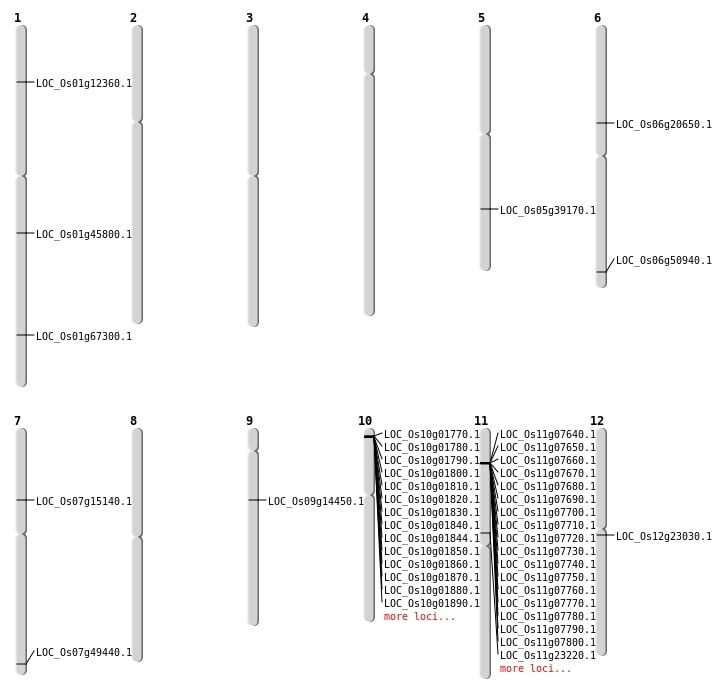
|
| Variant Information |
|---|
Single base substitutions: 57
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10642982 | C-T | Homo | ||
| SBS | Chr1 | 11894571 | G-A | HET | ||
| SBS | Chr1 | 20470322 | T-A | HET | ||
| SBS | Chr1 | 21659948 | C-A | HET | ||
| SBS | Chr1 | 26018832 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g45800 |
| SBS | Chr1 | 39064069 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g67300 |
| SBS | Chr1 | 41141646 | C-A | HET | ||
| SBS | Chr1 | 4179380 | T-A | HET | ||
| SBS | Chr1 | 4179381 | C-A | HET | ||
| SBS | Chr1 | 6754899 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g12360 |
| SBS | Chr1 | 9055591 | C-T | Homo | ||
| SBS | Chr10 | 10238505 | T-A | Homo | ||
| SBS | Chr10 | 1790951 | C-T | HET | ||
| SBS | Chr10 | 23098655 | A-G | HET | ||
| SBS | Chr10 | 3008738 | T-G | HET | ||
| SBS | Chr10 | 6064123 | T-C | Homo | ||
| SBS | Chr11 | 7622157 | C-A | HET | ||
| SBS | Chr11 | 8087822 | G-A | Homo | ||
| SBS | Chr11 | 8460332 | G-A | HET | ||
| SBS | Chr12 | 1013405 | G-A | Homo | ||
| SBS | Chr12 | 13019257 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g23030 |
| SBS | Chr12 | 13019258 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g23030 |
| SBS | Chr12 | 14289121 | G-C | HET | ||
| SBS | Chr12 | 15366433 | T-A | Homo | ||
| SBS | Chr12 | 23217907 | T-A | HET | ||
| SBS | Chr12 | 3167435 | T-C | HET | ||
| SBS | Chr12 | 672240 | C-A | Homo | ||
| SBS | Chr12 | 8930003 | G-A | HET | ||
| SBS | Chr2 | 10710907 | C-T | HET | ||
| SBS | Chr3 | 23450769 | C-A | Homo | ||
| SBS | Chr3 | 23450771 | T-A | Homo | ||
| SBS | Chr4 | 19277503 | C-T | HET | ||
| SBS | Chr4 | 19582073 | C-A | HET | ||
| SBS | Chr4 | 19800127 | C-A | HET | ||
| SBS | Chr4 | 20900321 | T-C | HET | ||
| SBS | Chr4 | 28550098 | A-T | HET | ||
| SBS | Chr4 | 30851457 | G-A | Homo | ||
| SBS | Chr5 | 22970976 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g39170 |
| SBS | Chr5 | 27305203 | G-A | HET | ||
| SBS | Chr5 | 29749055 | G-T | HET | ||
| SBS | Chr6 | 10820222 | C-T | HET | ||
| SBS | Chr6 | 11900520 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g20650 |
| SBS | Chr6 | 12872771 | G-A | HET | ||
| SBS | Chr6 | 15752740 | G-A | Homo | ||
| SBS | Chr6 | 21807973 | A-G | HET | ||
| SBS | Chr6 | 30828837 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g50940 |
| SBS | Chr6 | 3727378 | T-G | HET | ||
| SBS | Chr7 | 19151175 | A-T | HET | ||
| SBS | Chr7 | 19321840 | A-T | Homo | ||
| SBS | Chr7 | 24830382 | T-G | HET | ||
| SBS | Chr7 | 27480707 | G-A | HET | ||
| SBS | Chr7 | 29207589 | T-C | HET | ||
| SBS | Chr7 | 29606933 | G-A | HET | STOP_GAINED | LOC_Os07g49440 |
| SBS | Chr7 | 8692159 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g15140 |
| SBS | Chr7 | 9725195 | G-A | Homo | ||
| SBS | Chr8 | 8290374 | C-T | HET | ||
| SBS | Chr9 | 8539592 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14450 |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 49552 | 49554 | 2 | |
| Deletion | Chr10 | 505001 | 529000 | 23999 | 5 |
| Deletion | Chr10 | 532001 | 854000 | 321999 | 52 |
| Deletion | Chr12 | 672635 | 672637 | 2 | |
| Deletion | Chr12 | 868052 | 868095 | 43 | |
| Deletion | Chr8 | 2335306 | 2335307 | 1 | |
| Deletion | Chr11 | 3891001 | 3987000 | 95999 | 17 |
| Deletion | Chr4 | 6304476 | 6304487 | 11 | |
| Deletion | Chr10 | 6525679 | 6525699 | 20 | |
| Deletion | Chr5 | 7036438 | 7036441 | 3 | |
| Deletion | Chr10 | 10154662 | 10154664 | 2 | |
| Deletion | Chr3 | 11941012 | 11941018 | 6 | |
| Deletion | Chr4 | 12062449 | 12062451 | 2 | |
| Deletion | Chr12 | 12795281 | 12795293 | 12 | |
| Deletion | Chr11 | 14010961 | 14010974 | 13 | |
| Deletion | Chr12 | 15318424 | 15318435 | 11 | |
| Deletion | Chr11 | 16687294 | 16687295 | 1 | LOC_Os11g28790 |
| Deletion | Chr11 | 17065001 | 17077000 | 11999 | 2 |
| Deletion | Chr8 | 18992722 | 18992723 | 1 | |
| Deletion | Chr12 | 21780819 | 21780823 | 4 | |
| Deletion | Chr3 | 21858395 | 21858396 | 1 | |
| Deletion | Chr1 | 21903150 | 21903162 | 12 | |
| Deletion | Chr5 | 22953329 | 22953332 | 3 | |
| Deletion | Chr7 | 23832477 | 23832485 | 8 | |
| Deletion | Chr3 | 26540831 | 26540832 | 1 | |
| Deletion | Chr2 | 27237697 | 27237698 | 1 | |
| Deletion | Chr7 | 27955976 | 27955977 | 1 | |
| Deletion | Chr2 | 28431986 | 28431996 | 10 | |
| Deletion | Chr1 | 41047479 | 41047481 | 2 |
No Insertion
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 13348787 | 13378766 | 2 |
| Inversion | Chr11 | 13348805 | 13378774 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 16085015 | Chr1 | 10234283 | LOC_Os10g30850 |
