Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2698-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2698-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 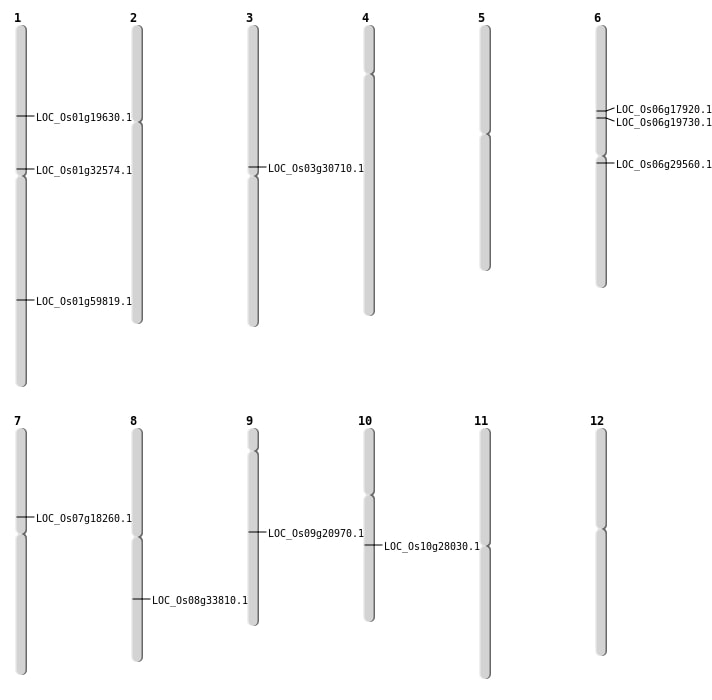
|
| Variant Information |
|---|
Single base substitutions: 56
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17869762 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g32574 |
| SBS | Chr1 | 32687294 | C-T | Homo | ||
| SBS | Chr1 | 32881072 | T-C | Homo | ||
| SBS | Chr10 | 14548165 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g28030 |
| SBS | Chr10 | 8250356 | C-T | HET | ||
| SBS | Chr11 | 12896705 | G-C | Homo | ||
| SBS | Chr11 | 18810787 | C-G | HET | ||
| SBS | Chr11 | 2937727 | T-A | HET | ||
| SBS | Chr11 | 9266171 | T-G | HET | ||
| SBS | Chr12 | 10632186 | C-T | HET | ||
| SBS | Chr12 | 14285489 | C-T | HET | ||
| SBS | Chr12 | 22546728 | T-A | HET | ||
| SBS | Chr2 | 10877203 | T-G | HET | ||
| SBS | Chr2 | 26403679 | T-C | HET | ||
| SBS | Chr2 | 27638002 | C-A | Homo | ||
| SBS | Chr2 | 3579220 | G-A | Homo | ||
| SBS | Chr3 | 17095852 | A-G | HET | ||
| SBS | Chr3 | 31985830 | G-A | HET | ||
| SBS | Chr3 | 6442551 | A-G | HET | ||
| SBS | Chr3 | 8627041 | G-A | HET | ||
| SBS | Chr3 | 8627042 | A-T | HET | ||
| SBS | Chr3 | 9614084 | C-T | HET | ||
| SBS | Chr4 | 10191396 | C-A | HET | ||
| SBS | Chr4 | 13459865 | A-G | HET | ||
| SBS | Chr4 | 15996126 | G-A | HET | ||
| SBS | Chr4 | 2374177 | G-T | HET | ||
| SBS | Chr4 | 26445094 | A-G | HET | ||
| SBS | Chr4 | 28713943 | T-C | Homo | ||
| SBS | Chr4 | 4883507 | C-G | HET | ||
| SBS | Chr5 | 1896467 | T-A | HET | ||
| SBS | Chr5 | 19106505 | C-T | HET | ||
| SBS | Chr5 | 1919616 | T-A | HET | ||
| SBS | Chr5 | 19876312 | G-A | HET | ||
| SBS | Chr5 | 23090816 | C-T | HET | ||
| SBS | Chr5 | 26122917 | A-T | HET | ||
| SBS | Chr5 | 26122918 | G-A | HET | ||
| SBS | Chr6 | 11280955 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g19730 |
| SBS | Chr6 | 16933699 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g29560 |
| SBS | Chr6 | 27144072 | A-T | HET | ||
| SBS | Chr7 | 12315401 | C-T | HET | ||
| SBS | Chr7 | 2654205 | T-C | HET | ||
| SBS | Chr7 | 26854960 | G-T | HET | ||
| SBS | Chr7 | 4283218 | G-A | HET | ||
| SBS | Chr7 | 5133398 | T-C | HET | ||
| SBS | Chr8 | 10993651 | G-A | Homo | ||
| SBS | Chr8 | 19513430 | G-T | HET | ||
| SBS | Chr8 | 3324647 | C-T | HET | ||
| SBS | Chr8 | 4115814 | T-C | HET | ||
| SBS | Chr8 | 6668657 | T-A | HET | ||
| SBS | Chr9 | 10214908 | A-G | HET | ||
| SBS | Chr9 | 12629607 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g20970 |
| SBS | Chr9 | 16141239 | A-G | HET | ||
| SBS | Chr9 | 21627895 | G-T | HET | ||
| SBS | Chr9 | 21923468 | C-A | HET | ||
| SBS | Chr9 | 3528595 | G-A | HET | ||
| SBS | Chr9 | 6342171 | A-C | HET |
Deletions: 26
Insertions: 7
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 10412514 | 10412739 | LOC_Os06g17920 |
| Inversion | Chr1 | 11130501 | 11130705 | LOC_Os01g19630 |
| Inversion | Chr8 | 21169429 | 21169539 | LOC_Os08g33810 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 754802 | Chr2 | 4019117 | |
| Translocation | Chr12 | 8980230 | Chr7 | 10825617 | 2 |
| Translocation | Chr6 | 13904216 | Chr1 | 34601118 | 2 |
| Translocation | Chr3 | 17497157 | Chr2 | 29156147 | LOC_Os03g30710 |
