Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2740-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2740-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 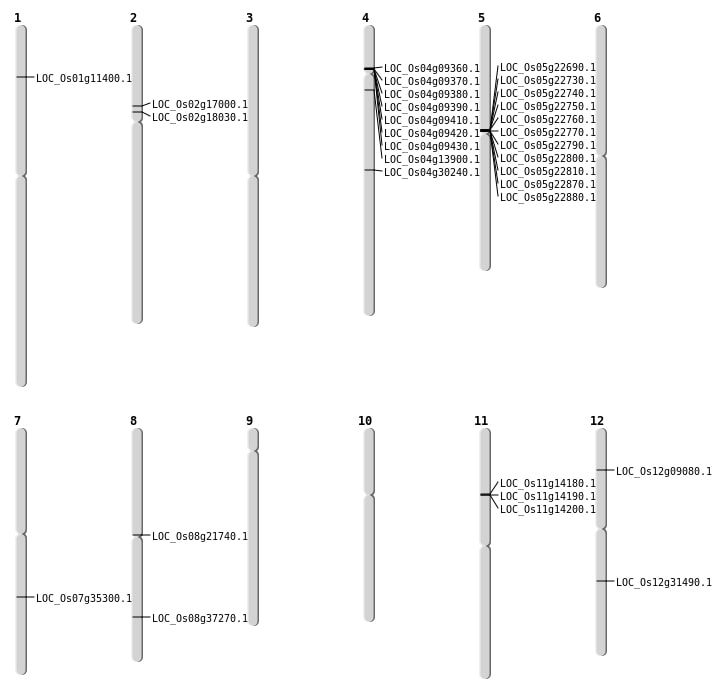
|
| Variant Information |
|---|
Single base substitutions: 53
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11424630 | T-C | HET | ||
| SBS | Chr1 | 2219671 | T-C | Homo | ||
| SBS | Chr1 | 31239916 | G-A | HET | ||
| SBS | Chr1 | 34111842 | A-C | HET | ||
| SBS | Chr1 | 36384513 | T-C | HET | ||
| SBS | Chr1 | 4448414 | C-T | HET | ||
| SBS | Chr1 | 6132442 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g11400 |
| SBS | Chr11 | 10771596 | C-T | HET | ||
| SBS | Chr12 | 10762243 | C-A | Homo | ||
| SBS | Chr12 | 12518705 | C-T | HET | ||
| SBS | Chr12 | 18941730 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g31490 |
| SBS | Chr12 | 27162429 | T-A | Homo | ||
| SBS | Chr12 | 4747153 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g09080 |
| SBS | Chr12 | 5529807 | A-T | Homo | ||
| SBS | Chr2 | 13932938 | G-A | HET | ||
| SBS | Chr2 | 14519563 | C-T | HET | ||
| SBS | Chr2 | 20238549 | C-G | HET | ||
| SBS | Chr2 | 30164250 | G-A | HET | ||
| SBS | Chr2 | 30164251 | G-A | HET | ||
| SBS | Chr2 | 9715903 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g17000 |
| SBS | Chr3 | 19033161 | T-C | HET | ||
| SBS | Chr3 | 21819939 | T-A | HET | ||
| SBS | Chr3 | 21819945 | A-T | HET | ||
| SBS | Chr3 | 25228833 | C-T | HET | ||
| SBS | Chr3 | 30873355 | T-C | HET | ||
| SBS | Chr3 | 8374629 | T-C | HET | ||
| SBS | Chr4 | 11227539 | A-G | HET | ||
| SBS | Chr4 | 14118574 | A-T | HET | ||
| SBS | Chr4 | 18043871 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g30240 |
| SBS | Chr4 | 6826323 | G-T | HET | ||
| SBS | Chr4 | 7762984 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g13900 |
| SBS | Chr4 | 9476105 | G-A | HET | ||
| SBS | Chr4 | 9860881 | C-G | HET | ||
| SBS | Chr5 | 13397220 | C-T | HET | ||
| SBS | Chr5 | 16791259 | G-A | HET | ||
| SBS | Chr5 | 19400239 | C-A | HET | ||
| SBS | Chr5 | 19425955 | C-A | HET | ||
| SBS | Chr5 | 21177289 | C-T | HET | ||
| SBS | Chr5 | 4042721 | T-A | HET | ||
| SBS | Chr6 | 12030120 | C-A | HET | ||
| SBS | Chr6 | 19739126 | T-G | HET | ||
| SBS | Chr6 | 22177702 | G-A | Homo | ||
| SBS | Chr6 | 5434705 | G-T | Homo | ||
| SBS | Chr6 | 7456794 | T-A | HET | ||
| SBS | Chr7 | 17490713 | A-G | HET | ||
| SBS | Chr7 | 1845655 | A-C | HET | ||
| SBS | Chr7 | 21592260 | T-C | HET | ||
| SBS | Chr7 | 8070534 | G-A | HET | ||
| SBS | Chr8 | 13023237 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g21740 |
| SBS | Chr8 | 16269214 | G-T | Homo | ||
| SBS | Chr8 | 7666358 | T-C | Homo | ||
| SBS | Chr9 | 1905449 | C-T | HET | ||
| SBS | Chr9 | 3903748 | T-G | HET |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 701702 | 701703 | 1 | |
| Deletion | Chr2 | 1530441 | 1530449 | 8 | |
| Deletion | Chr4 | 4980001 | 5046000 | 65999 | 7 |
| Deletion | Chr4 | 5999872 | 5999880 | 8 | |
| Deletion | Chr11 | 7920001 | 7948000 | 27999 | 3 |
| Deletion | Chr6 | 10900133 | 10900138 | 5 | |
| Deletion | Chr10 | 11257398 | 11257401 | 3 | |
| Deletion | Chr10 | 11271297 | 11271299 | 2 | |
| Deletion | Chr5 | 12892001 | 12947000 | 54999 | 8 |
| Deletion | Chr5 | 12990001 | 13006000 | 15999 | 2 |
| Deletion | Chr6 | 17390465 | 17390466 | 1 | |
| Deletion | Chr12 | 18864599 | 18864605 | 6 | |
| Deletion | Chr6 | 20105934 | 20105941 | 7 | |
| Deletion | Chr2 | 20585150 | 20585152 | 2 | |
| Deletion | Chr7 | 21129096 | 21129099 | 3 | LOC_Os07g35300 |
| Deletion | Chr4 | 21198079 | 21198199 | 120 | |
| Deletion | Chr8 | 22949001 | 22968000 | 18999 | |
| Deletion | Chr5 | 23570870 | 23570872 | 2 | |
| Deletion | Chr3 | 23878924 | 23878930 | 6 | |
| Deletion | Chr8 | 26360943 | 26360944 | 1 | |
| Deletion | Chr1 | 34298081 | 34298082 | 1 | |
| Deletion | Chr1 | 39256953 | 39256958 | 5 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 19500625 | 19500630 | 6 | |
| Insertion | Chr5 | 1631783 | 1631783 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 7921714 | 8852447 | |
| Inversion | Chr11 | 7947947 | 8852451 | |
| Inversion | Chr8 | 22948499 | 23558334 | 2 |
| Inversion | Chr8 | 22967889 | 23558338 | 2 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 9487773 | Chr6 | 12598603 | |
| Translocation | Chr11 | 9960751 | Chr10 | 18647685 | |
| Translocation | Chr5 | 12880759 | Chr3 | 24381807 | LOC_Os05g22690 |
| Translocation | Chr5 | 13006027 | Chr3 | 24376609 | |
| Translocation | Chr10 | 15800904 | Chr4 | 13170888 | |
| Translocation | Chr9 | 16680084 | Chr2 | 10459219 | 2 |
| Translocation | Chr10 | 18722714 | Chr4 | 5043775 |
