Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2753-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2753-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 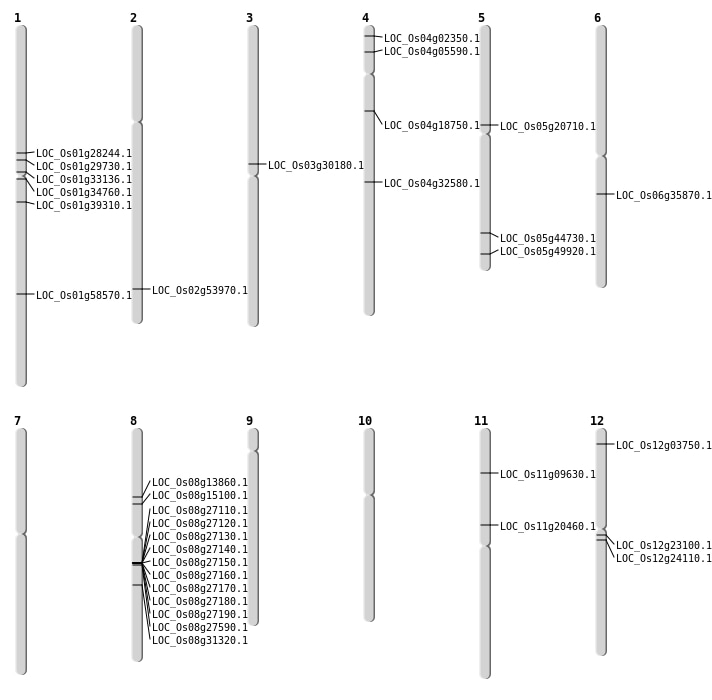
|
| Variant Information |
|---|
Single base substitutions: 113
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12999526 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 15824176 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g28244 |
| SBS | Chr1 | 16657453 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g29730 |
| SBS | Chr1 | 17714943 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 17905812 | G-A | HET | ||
| SBS | Chr1 | 18241368 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g33136 |
| SBS | Chr1 | 19869737 | C-T | HET | ||
| SBS | Chr1 | 22126670 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g39310 |
| SBS | Chr1 | 37745648 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr10 | 2692699 | A-G | HET | ||
| SBS | Chr10 | 6468134 | T-A | HET | ||
| SBS | Chr10 | 6488320 | G-A | HET | ||
| SBS | Chr11 | 13463650 | C-A | HOMO | INTRON | |
| SBS | Chr11 | 15050766 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18980858 | T-A | HET | ||
| SBS | Chr11 | 22125403 | G-T | HET | ||
| SBS | Chr11 | 25338030 | C-T | HET | ||
| SBS | Chr11 | 3124308 | G-T | Homo | ||
| SBS | Chr11 | 5149599 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g09630 |
| SBS | Chr11 | 5794257 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 8638112 | A-C | HET | INTRON | |
| SBS | Chr12 | 10670185 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr12 | 10957923 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 11441647 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 11973232 | G-T | HET | ||
| SBS | Chr12 | 13064266 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g23100 |
| SBS | Chr12 | 13719698 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g24110 |
| SBS | Chr12 | 19341322 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 19343331 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 20945927 | T-A | Homo | ||
| SBS | Chr12 | 21276986 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 5291350 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 8373141 | C-G | HET | ||
| SBS | Chr12 | 9883563 | G-A | HET | ||
| SBS | Chr2 | 13981702 | G-C | HET | ||
| SBS | Chr2 | 21073994 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 2138606 | G-A | HET | ||
| SBS | Chr2 | 21978201 | G-A | Homo | ||
| SBS | Chr2 | 22166422 | C-T | HET | ||
| SBS | Chr2 | 28020379 | A-T | HET | ||
| SBS | Chr2 | 29695555 | C-T | HET | INTRON | |
| SBS | Chr2 | 33047059 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g53970 |
| SBS | Chr2 | 33078448 | C-G | HET | START_GAINED | |
| SBS | Chr2 | 33078452 | G-T | HET | UTR_5_PRIME | |
| SBS | Chr2 | 33674453 | A-T | HET | INTRON | |
| SBS | Chr3 | 11016643 | G-A | Homo | ||
| SBS | Chr3 | 17224578 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g30180 |
| SBS | Chr3 | 19807278 | C-T | Homo | ||
| SBS | Chr3 | 20500872 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 28800928 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 28881227 | C-A | HET | ||
| SBS | Chr3 | 29509764 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 30058467 | G-A | HET | ||
| SBS | Chr3 | 31281611 | C-T | HET | ||
| SBS | Chr3 | 32175826 | G-A | HET | INTRON | |
| SBS | Chr3 | 34375481 | G-C | HET | ||
| SBS | Chr3 | 7682798 | A-T | HET | ||
| SBS | Chr4 | 10403966 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18750 |
| SBS | Chr4 | 11957221 | C-T | HET | ||
| SBS | Chr4 | 13261573 | G-A | Homo | ||
| SBS | Chr4 | 14794999 | T-G | Homo | ||
| SBS | Chr4 | 18127627 | G-A | HET | ||
| SBS | Chr4 | 18883696 | C-A | Homo | ||
| SBS | Chr4 | 18883697 | T-C | Homo | ||
| SBS | Chr4 | 19214358 | T-C | Homo | ||
| SBS | Chr4 | 21204477 | C-A | Homo | ||
| SBS | Chr4 | 2844729 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g05590 |
| SBS | Chr4 | 9449393 | C-G | HET | ||
| SBS | Chr5 | 11762075 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 12140692 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g20710 |
| SBS | Chr5 | 13966379 | A-G | Homo | ||
| SBS | Chr5 | 22972903 | C-T | HET | ||
| SBS | Chr5 | 23531970 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 25534120 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 26333603 | T-C | Homo | ||
| SBS | Chr5 | 27546792 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 28633800 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g49920 |
| SBS | Chr5 | 6736152 | T-A | Homo | ||
| SBS | Chr5 | 9070881 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 9070882 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 11963168 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 13295840 | T-A | Homo | ||
| SBS | Chr6 | 26588652 | A-G | HET | ||
| SBS | Chr6 | 27984388 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 29084919 | G-A | HET | ||
| SBS | Chr6 | 29479680 | T-C | HET | ||
| SBS | Chr6 | 3336626 | G-A | Homo | ||
| SBS | Chr7 | 14862611 | C-T | Homo | ||
| SBS | Chr7 | 17135547 | G-A | HET | ||
| SBS | Chr7 | 27503070 | T-C | HET | INTRON | |
| SBS | Chr7 | 29425169 | C-A | HET | ||
| SBS | Chr7 | 399401 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 8724830 | G-A | HET | ||
| SBS | Chr8 | 1157317 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 12461122 | G-A | HET | INTRON | |
| SBS | Chr8 | 13660220 | C-T | HET | ||
| SBS | Chr8 | 19597116 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 20721027 | G-A | HET | INTRON | |
| SBS | Chr8 | 2199926 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 3384582 | A-G | HET | ||
| SBS | Chr8 | 446736 | G-A | HET | ||
| SBS | Chr8 | 7049914 | G-A | Homo | ||
| SBS | Chr8 | 8276829 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g13860 |
| SBS | Chr8 | 9151688 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g15100 |
| SBS | Chr9 | 11207196 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 12047368 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 12306681 | G-A | HET | INTRON | |
| SBS | Chr9 | 19362809 | A-G | HET | ||
| SBS | Chr9 | 19400857 | C-T | HET | ||
| SBS | Chr9 | 20518934 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 20825910 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 5992744 | G-T | HET | INTRON | |
| SBS | Chr9 | 7499894 | G-C | HET | INTERGENIC |
Deletions: 41
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 833506 | 833507 | 1 | LOC_Os04g02350 |
| Deletion | Chr5 | 1327551 | 1327591 | 40 | |
| Deletion | Chr12 | 1524590 | 1524596 | 6 | LOC_Os12g03750 |
| Deletion | Chr7 | 1915834 | 1915837 | 3 | |
| Deletion | Chr9 | 2225531 | 2225601 | 70 | |
| Deletion | Chr8 | 2543625 | 2543688 | 63 | |
| Deletion | Chr4 | 3510044 | 3510045 | 1 | |
| Deletion | Chr4 | 5349104 | 5349130 | 26 | |
| Deletion | Chr5 | 6411381 | 6411382 | 1 | |
| Deletion | Chr12 | 6826381 | 6826382 | 1 | |
| Deletion | Chr6 | 7240174 | 7240179 | 5 | |
| Deletion | Chr12 | 8797140 | 8797142 | 2 | 2 |
| Deletion | Chr7 | 8851707 | 8851751 | 44 | |
| Deletion | Chr6 | 10037983 | 10037987 | 4 | |
| Deletion | Chr7 | 10968349 | 10968350 | 1 | |
| Deletion | Chr6 | 11485476 | 11485487 | 11 | |
| Deletion | Chr7 | 12767245 | 12767247 | 2 | |
| Deletion | Chr4 | 13010166 | 13010170 | 4 | |
| Deletion | Chr5 | 13541735 | 13541769 | 34 | |
| Deletion | Chr7 | 13579697 | 13579698 | 1 | |
| Deletion | Chr4 | 14753982 | 14753989 | 7 | |
| Deletion | Chr8 | 16556001 | 16576000 | 19999 | 3 |
| Deletion | Chr8 | 16579001 | 16611000 | 31999 | 7 |
| Deletion | Chr11 | 18956067 | 18956068 | 1 | |
| Deletion | Chr9 | 20666289 | 20666290 | 1 | |
| Deletion | Chr12 | 20910937 | 20910938 | 1 | |
| Deletion | Chr3 | 21439707 | 21439722 | 15 | |
| Deletion | Chr8 | 21734592 | 21734594 | 2 | |
| Deletion | Chr2 | 21926504 | 21926507 | 3 | |
| Deletion | Chr4 | 22303876 | 22303878 | 2 | |
| Deletion | Chr8 | 24724154 | 24724156 | 2 | |
| Deletion | Chr1 | 27205030 | 27205033 | 3 | |
| Deletion | Chr12 | 27205500 | 27205505 | 5 | |
| Deletion | Chr5 | 27665648 | 27665664 | 16 | |
| Deletion | Chr4 | 28095714 | 28095742 | 28 | |
| Deletion | Chr6 | 29334764 | 29334774 | 10 | |
| Deletion | Chr4 | 30324190 | 30324202 | 12 | |
| Deletion | Chr3 | 31121859 | 31121863 | 4 | |
| Deletion | Chr3 | 36045962 | 36045964 | 2 | |
| Deletion | Chr1 | 39835952 | 39835955 | 3 | |
| Deletion | Chr1 | 42653212 | 42653218 | 6 |
Insertions: 14
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 1373374 | 1402214 | |
| Inversion | Chr12 | 1373378 | 1402219 | |
| Inversion | Chr11 | 11660550 | 11660943 | |
| Inversion | Chr2 | 18582765 | 18583272 | |
| Inversion | Chr4 | 19612218 | 19612385 | LOC_Os04g32580 |
Translocations: 19
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 3766431 | Chr11 | 17795284 | |
| Translocation | Chr12 | 4788930 | Chr3 | 16568136 | |
| Translocation | Chr11 | 5733929 | Chr1 | 10234167 | |
| Translocation | Chr10 | 8110736 | Chr1 | 16916018 | |
| Translocation | Chr12 | 8582249 | Chr1 | 19169551 | LOC_Os01g34760 |
| Translocation | Chr11 | 10659256 | Chr6 | 15618050 | |
| Translocation | Chr8 | 12890399 | Chr5 | 26582797 | |
| Translocation | Chr8 | 12890406 | Chr1 | 20080061 | |
| Translocation | Chr12 | 13923178 | Chr2 | 19373260 | |
| Translocation | Chr12 | 14379827 | Chr11 | 11845209 | LOC_Os11g20460 |
| Translocation | Chr12 | 15191193 | Chr4 | 18885976 | |
| Translocation | Chr8 | 19371940 | Chr7 | 20951359 | LOC_Os08g31320 |
| Translocation | Chr8 | 19372459 | Chr7 | 20951362 | |
| Translocation | Chr8 | 20584384 | Chr6 | 1072417 | |
| Translocation | Chr6 | 20931162 | Chr1 | 22551268 | LOC_Os06g35870 |
| Translocation | Chr9 | 21260004 | Chr8 | 24138969 | |
| Translocation | Chr11 | 22608900 | Chr9 | 11816282 | |
| Translocation | Chr5 | 26005965 | Chr1 | 33854332 | 2 |
| Translocation | Chr11 | 27758202 | Chr4 | 3130962 |
