Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2755-S-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2755-S-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 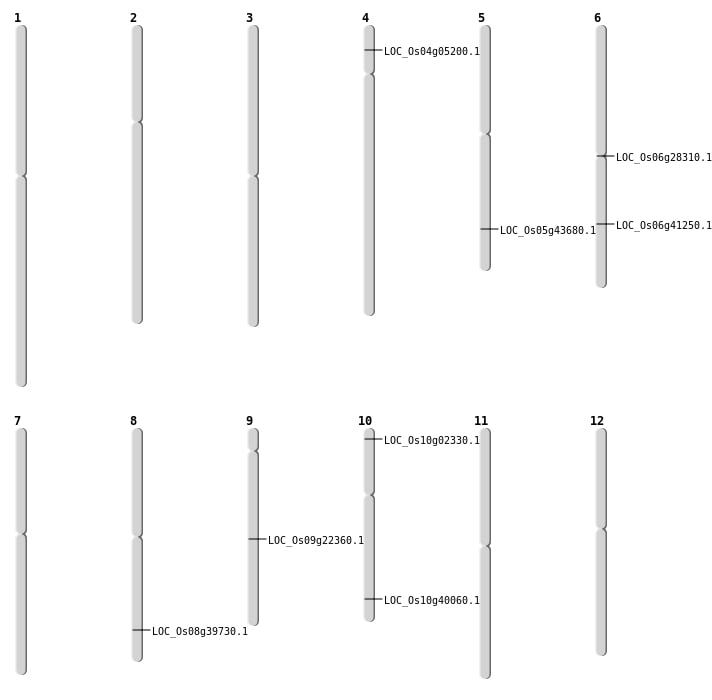
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 37507648 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 37507649 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 9434029 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr10 | 17730796 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3642139 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 4405639 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16284265 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 20841243 | A-C | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 2634256 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 32813563 | C-T | HET | INTRON | |
| SBS | Chr2 | 33538703 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 8657231 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 23504046 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 26889953 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 2596474 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g05200 |
| SBS | Chr4 | 33051752 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 3396544 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 7640675 | A-T | HOMO | INTRON | |
| SBS | Chr6 | 25372163 | A-T | HET | INTRON | |
| SBS | Chr6 | 2740088 | C-T | HET | INTRON | |
| SBS | Chr7 | 14338951 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 2042434 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 20923497 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 2524882 | G-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 25156066 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g39730 |
| SBS | Chr8 | 3411296 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 13514650 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g22360 |
| SBS | Chr9 | 19119158 | G-A | HET | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 64870 | 65224 | 354 | |
| Deletion | Chr7 | 1040851 | 1040856 | 5 | |
| Deletion | Chr6 | 1706333 | 1706335 | 2 | |
| Deletion | Chr7 | 6227649 | 6227652 | 3 | |
| Deletion | Chr12 | 8068786 | 8068788 | 2 | |
| Deletion | Chr4 | 11041854 | 11041856 | 2 | |
| Deletion | Chr12 | 11725122 | 11725136 | 14 | |
| Deletion | Chr4 | 13754369 | 13754370 | 1 | |
| Deletion | Chr4 | 14868171 | 14868172 | 1 | |
| Deletion | Chr6 | 16082829 | 16082830 | 1 | LOC_Os06g28310 |
| Deletion | Chr6 | 16785006 | 16785011 | 5 | |
| Deletion | Chr5 | 18350712 | 18350713 | 1 | |
| Deletion | Chr1 | 19947915 | 19947916 | 1 | |
| Deletion | Chr5 | 20510461 | 20510462 | 1 | |
| Deletion | Chr10 | 21456586 | 21456591 | 5 | LOC_Os10g40060 |
| Deletion | Chr2 | 22470466 | 22470474 | 8 | |
| Deletion | Chr11 | 25389394 | 25389396 | 2 | |
| Deletion | Chr5 | 25393618 | 25393627 | 9 | LOC_Os05g43680 |
| Deletion | Chr3 | 27678245 | 27678246 | 1 |
Insertions: 21
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 833387 | 833735 | LOC_Os10g02330 |
| Inversion | Chr11 | 18825586 | 18828949 | |
| Inversion | Chr6 | 24684474 | 24686396 | LOC_Os06g41250 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 1617625 | Chr1 | 18222002 | |
| Translocation | Chr5 | 5657243 | Chr4 | 27176915 | |
| Translocation | Chr8 | 16449095 | Chr6 | 24693727 | |
| Translocation | Chr11 | 19361833 | Chr2 | 7462906 | |
| Translocation | Chr11 | 19361845 | Chr2 | 7462449 |
