Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2782-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2782-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 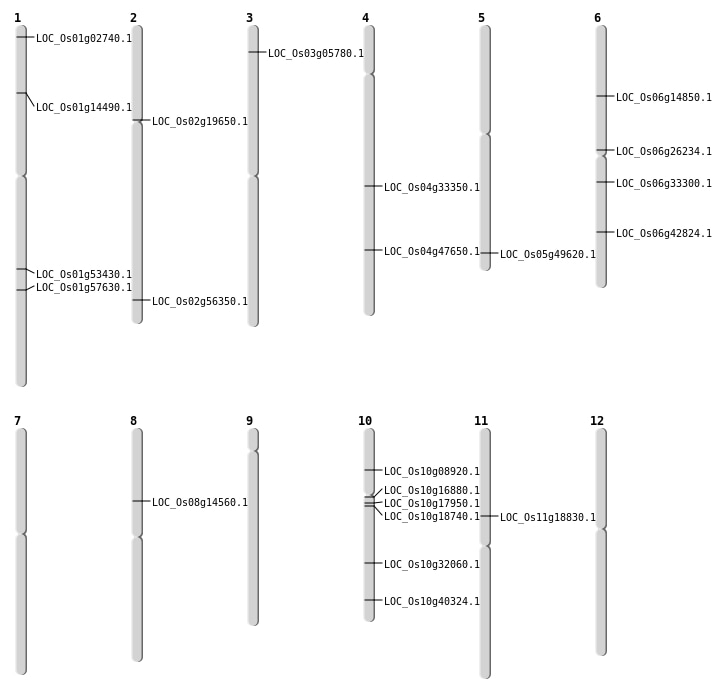
|
| Variant Information |
|---|
Single base substitutions: 53
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 35777155 | T-G | HET | ||
| SBS | Chr1 | 5372320 | A-T | HET | ||
| SBS | Chr1 | 7077226 | T-A | HET | ||
| SBS | Chr10 | 12429467 | A-T | HET | ||
| SBS | Chr10 | 1261342 | G-C | HET | ||
| SBS | Chr10 | 20628177 | G-T | HET | ||
| SBS | Chr10 | 2437289 | C-T | HET | ||
| SBS | Chr10 | 4827129 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g08920 |
| SBS | Chr10 | 8412596 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g16880 |
| SBS | Chr10 | 9088569 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g17950 |
| SBS | Chr10 | 9531671 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g18740 |
| SBS | Chr11 | 11974037 | C-T | HET | ||
| SBS | Chr11 | 17860464 | G-A | HET | ||
| SBS | Chr11 | 18062055 | G-C | HET | ||
| SBS | Chr11 | 9837952 | G-A | HET | ||
| SBS | Chr12 | 11079043 | C-T | HET | ||
| SBS | Chr12 | 17683833 | G-A | HET | ||
| SBS | Chr12 | 26114854 | C-A | HET | ||
| SBS | Chr12 | 2838047 | G-A | HET | ||
| SBS | Chr2 | 11330899 | C-T | HET | ||
| SBS | Chr2 | 26350002 | T-C | HET | ||
| SBS | Chr2 | 34492717 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g56350 |
| SBS | Chr2 | 34768141 | G-C | HET | ||
| SBS | Chr2 | 6358439 | C-T | HET | ||
| SBS | Chr2 | 7831349 | C-G | HET | ||
| SBS | Chr3 | 10308797 | T-A | HET | ||
| SBS | Chr3 | 21766880 | G-T | HET | ||
| SBS | Chr3 | 33496738 | C-T | HET | ||
| SBS | Chr4 | 20195814 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g33350 |
| SBS | Chr4 | 23724615 | G-A | HET | ||
| SBS | Chr4 | 7096917 | C-G | HET | ||
| SBS | Chr4 | 7181104 | C-T | HET | ||
| SBS | Chr4 | 9477843 | C-A | HET | ||
| SBS | Chr5 | 11297071 | T-C | Homo | ||
| SBS | Chr5 | 22810958 | A-G | HET | ||
| SBS | Chr5 | 28160403 | G-T | HET | ||
| SBS | Chr5 | 28472772 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g49620 |
| SBS | Chr6 | 10698379 | C-T | HET | ||
| SBS | Chr6 | 27912934 | G-A | HET | ||
| SBS | Chr6 | 28787460 | G-A | Homo | ||
| SBS | Chr7 | 12309407 | C-T | HET | ||
| SBS | Chr7 | 15035128 | G-A | HET | ||
| SBS | Chr7 | 15385328 | T-C | HET | ||
| SBS | Chr7 | 15577405 | A-T | HET | ||
| SBS | Chr7 | 20353357 | A-G | HET | ||
| SBS | Chr7 | 21240030 | C-T | HET | ||
| SBS | Chr7 | 26867085 | C-T | HET | ||
| SBS | Chr8 | 16242865 | A-T | HET | ||
| SBS | Chr8 | 17681852 | C-G | HET | ||
| SBS | Chr8 | 18391490 | G-C | Homo | ||
| SBS | Chr8 | 19742921 | T-C | HET | ||
| SBS | Chr8 | 5062470 | C-T | HET | ||
| SBS | Chr8 | 8753603 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g14560 |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 591392 | 591400 | 8 | |
| Deletion | Chr12 | 933460 | 933462 | 2 | |
| Deletion | Chr1 | 943420 | 943425 | 5 | LOC_Os01g02740 |
| Deletion | Chr9 | 2398191 | 2398195 | 4 | |
| Deletion | Chr3 | 2884824 | 2884825 | 1 | LOC_Os03g05780 |
| Deletion | Chr1 | 7962537 | 7962544 | 7 | |
| Deletion | Chr1 | 8117645 | 8117657 | 12 | LOC_Os01g14490 |
| Deletion | Chr6 | 8445773 | 8445784 | 11 | LOC_Os06g14850 |
| Deletion | Chr12 | 9651718 | 9651722 | 4 | |
| Deletion | Chr3 | 11310152 | 11310186 | 34 | |
| Deletion | Chr2 | 11485486 | 11485489 | 3 | LOC_Os02g19650 |
| Deletion | Chr6 | 15350865 | 15350868 | 3 | LOC_Os06g26234 |
| Deletion | Chr2 | 15357615 | 15357628 | 13 | |
| Deletion | Chr10 | 16842299 | 16842304 | 5 | LOC_Os10g32060 |
| Deletion | Chr10 | 19471199 | 19471207 | 8 | |
| Deletion | Chr8 | 19860654 | 19860662 | 8 | |
| Deletion | Chr7 | 20872822 | 20872823 | 1 | |
| Deletion | Chr4 | 21905216 | 21905217 | 1 | |
| Deletion | Chr5 | 22369446 | 22369450 | 4 | |
| Deletion | Chr1 | 22680227 | 22680228 | 1 | |
| Deletion | Chr5 | 22852033 | 22852034 | 1 | |
| Deletion | Chr7 | 25254692 | 25254693 | 1 | |
| Deletion | Chr8 | 26352924 | 26352925 | 1 | |
| Deletion | Chr2 | 27443710 | 27443712 | 2 | |
| Deletion | Chr4 | 28278665 | 28278880 | 215 | LOC_Os04g47650 |
| Deletion | Chr1 | 30696571 | 30696580 | 9 | LOC_Os01g53430 |
| Deletion | Chr3 | 32641582 | 32641584 | 2 | |
| Deletion | Chr1 | 33318691 | 33318696 | 5 | LOC_Os01g57630 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 17442466 | 17442470 | 5 | |
| Insertion | Chr6 | 19387623 | 19387623 | 1 | LOC_Os06g33300 |
| Insertion | Chr7 | 1308318 | 1308335 | 18 | |
| Insertion | Chr9 | 8917715 | 8917717 | 3 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 10695745 | 11254629 | LOC_Os11g18830 |
| Inversion | Chr11 | 10695749 | 11254647 | LOC_Os11g18830 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 21612530 | Chr6 | 25743074 | 2 |
