Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2785-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2785-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 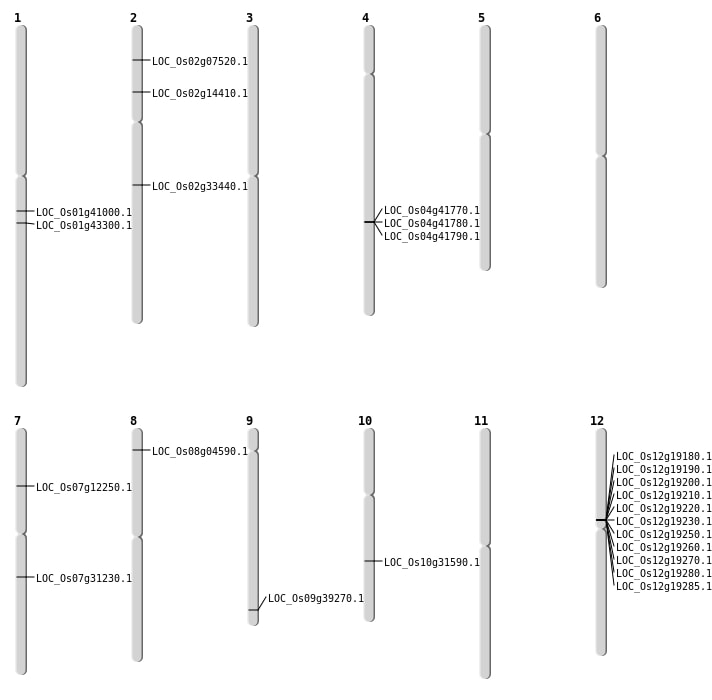
|
| Variant Information |
|---|
Single base substitutions: 55
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1509169 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g03660 |
| SBS | Chr1 | 23214690 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g41000 |
| SBS | Chr1 | 24744468 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g43300 |
| SBS | Chr1 | 42013764 | T-C | HET | ||
| SBS | Chr1 | 7617379 | A-G | HET | ||
| SBS | Chr10 | 14317354 | C-T | HET | ||
| SBS | Chr10 | 16555521 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g31590 |
| SBS | Chr10 | 2731554 | G-C | HET | ||
| SBS | Chr10 | 7330545 | C-G | HET | ||
| SBS | Chr11 | 12897101 | G-A | HET | ||
| SBS | Chr11 | 14569501 | C-T | HET | ||
| SBS | Chr11 | 14573368 | C-T | HET | ||
| SBS | Chr11 | 21087054 | A-C | HET | ||
| SBS | Chr11 | 21201812 | A-C | HET | ||
| SBS | Chr11 | 26845765 | C-T | HET | ||
| SBS | Chr11 | 7761165 | G-A | HET | ||
| SBS | Chr11 | 9678464 | A-G | HET | ||
| SBS | Chr12 | 16675240 | G-T | HET | ||
| SBS | Chr12 | 19828183 | T-A | HET | ||
| SBS | Chr2 | 1261592 | A-C | HET | ||
| SBS | Chr2 | 13241345 | T-A | HET | ||
| SBS | Chr2 | 13241346 | C-A | HET | ||
| SBS | Chr2 | 15990259 | T-A | HET | ||
| SBS | Chr2 | 19878326 | C-A | HET | STOP_GAINED | LOC_Os02g33440 |
| SBS | Chr2 | 22780769 | G-A | HET | ||
| SBS | Chr2 | 32847064 | G-A | HET | ||
| SBS | Chr2 | 34555016 | A-G | HET | ||
| SBS | Chr2 | 3892698 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g07520 |
| SBS | Chr2 | 7913172 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g14410 |
| SBS | Chr3 | 10344021 | G-T | HET | ||
| SBS | Chr3 | 13328022 | T-A | HET | ||
| SBS | Chr3 | 21716990 | C-T | HET | ||
| SBS | Chr3 | 21716991 | C-T | HET | ||
| SBS | Chr3 | 31311879 | G-T | HET | ||
| SBS | Chr4 | 29050025 | C-A | HET | ||
| SBS | Chr4 | 29247289 | G-T | HET | ||
| SBS | Chr4 | 9626315 | G-A | Homo | ||
| SBS | Chr5 | 17472787 | G-A | HET | ||
| SBS | Chr6 | 14631535 | G-A | Homo | ||
| SBS | Chr6 | 15688165 | C-A | HET | ||
| SBS | Chr6 | 16917794 | T-A | Homo | ||
| SBS | Chr6 | 24681512 | G-T | HET | ||
| SBS | Chr6 | 25473828 | C-T | HET | ||
| SBS | Chr6 | 29973101 | C-T | HET | ||
| SBS | Chr6 | 3227090 | T-A | HET | ||
| SBS | Chr7 | 16732701 | G-A | HET | ||
| SBS | Chr7 | 18486799 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g31230 |
| SBS | Chr7 | 26835152 | G-A | HET | ||
| SBS | Chr7 | 6865699 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g12250 |
| SBS | Chr7 | 834391 | G-A | HET | ||
| SBS | Chr8 | 7109305 | C-A | HET | ||
| SBS | Chr8 | 820179 | A-C | HET | ||
| SBS | Chr9 | 11081648 | C-T | HET | ||
| SBS | Chr9 | 13237020 | G-A | HET | ||
| SBS | Chr9 | 365120 | G-A | HET |
Deletions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1633135 | 1633137 | 2 | |
| Deletion | Chr8 | 3197341 | 3197347 | 6 | |
| Deletion | Chr2 | 6586019 | 6586020 | 1 | |
| Deletion | Chr7 | 6971696 | 6971698 | 2 | |
| Deletion | Chr12 | 11137001 | 11184000 | 46999 | 11 |
| Deletion | Chr9 | 13719565 | 13719576 | 11 | |
| Deletion | Chr3 | 16039365 | 16039367 | 2 | |
| Deletion | Chr9 | 22568364 | 22568376 | 12 | LOC_Os09g39270 |
| Deletion | Chr3 | 23512723 | 23512724 | 1 | |
| Deletion | Chr4 | 24751001 | 24760000 | 8999 | 3 |
| Deletion | Chr1 | 29822129 | 29822130 | 1 | |
| Deletion | Chr1 | 42047097 | 42047098 | 1 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 12943218 | 12943218 | 1 |
Inversions: 5
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3680107 | Chr8 | 1215904 | |
| Translocation | Chr9 | 8398273 | Chr2 | 25035289 |
