Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2795-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2795-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 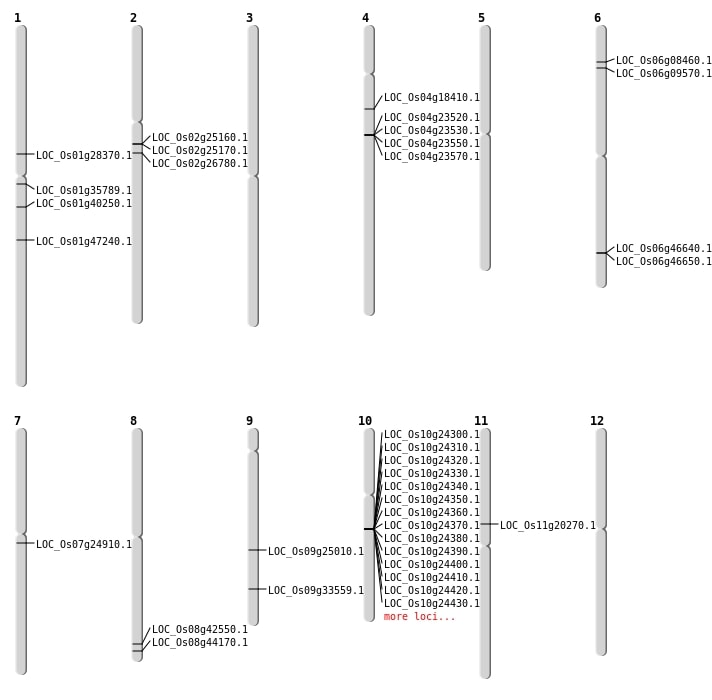
|
| Variant Information |
|---|
Single base substitutions: 65
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15804025 | T-A | HET | ||
| SBS | Chr1 | 15896428 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g28370 |
| SBS | Chr1 | 16241205 | G-T | HET | ||
| SBS | Chr1 | 16709369 | G-A | HET | ||
| SBS | Chr1 | 19448887 | G-T | HET | ||
| SBS | Chr1 | 22709212 | T-A | Homo | ||
| SBS | Chr1 | 30290364 | G-T | HET | ||
| SBS | Chr10 | 1468130 | A-C | HET | ||
| SBS | Chr10 | 3752696 | G-A | HET | ||
| SBS | Chr11 | 11979986 | G-T | HET | ||
| SBS | Chr11 | 20546670 | G-C | HET | ||
| SBS | Chr11 | 20998738 | G-T | HET | ||
| SBS | Chr11 | 22217528 | G-T | HET | ||
| SBS | Chr11 | 4374351 | C-T | HET | ||
| SBS | Chr11 | 4374352 | T-C | HET | ||
| SBS | Chr11 | 4416903 | A-G | HET | ||
| SBS | Chr11 | 9549510 | T-A | HET | ||
| SBS | Chr12 | 10251946 | G-T | HET | ||
| SBS | Chr12 | 14069421 | G-A | HET | ||
| SBS | Chr12 | 17642706 | C-T | HET | ||
| SBS | Chr12 | 24539957 | C-T | Homo | ||
| SBS | Chr2 | 10194831 | C-A | HET | ||
| SBS | Chr2 | 14039614 | G-T | HET | ||
| SBS | Chr2 | 2495545 | G-A | HET | ||
| SBS | Chr2 | 9914387 | G-A | HET | ||
| SBS | Chr3 | 25827741 | G-T | HET | ||
| SBS | Chr3 | 30920396 | T-A | HET | ||
| SBS | Chr4 | 10166406 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18410 |
| SBS | Chr4 | 13491223 | G-A | HET | ||
| SBS | Chr4 | 149339 | G-C | HET | ||
| SBS | Chr4 | 2146823 | G-A | HET | ||
| SBS | Chr4 | 24045950 | G-A | HET | ||
| SBS | Chr4 | 7565470 | G-T | HET | ||
| SBS | Chr4 | 8998768 | C-T | HET | ||
| SBS | Chr5 | 14025559 | A-T | Homo | ||
| SBS | Chr5 | 14025560 | G-T | Homo | ||
| SBS | Chr5 | 15400605 | G-A | HET | ||
| SBS | Chr5 | 16088362 | T-C | Homo | ||
| SBS | Chr5 | 24618049 | G-A | HET | ||
| SBS | Chr5 | 26917165 | A-T | HET | ||
| SBS | Chr5 | 27940643 | T-C | HET | ||
| SBS | Chr5 | 3177772 | G-T | HET | ||
| SBS | Chr5 | 4491165 | C-T | HET | ||
| SBS | Chr5 | 9881393 | A-T | HET | ||
| SBS | Chr5 | 9881394 | G-A | HET | ||
| SBS | Chr6 | 22288501 | G-T | Homo | ||
| SBS | Chr6 | 23658054 | G-T | HET | ||
| SBS | Chr6 | 2450003 | C-A | HET | ||
| SBS | Chr6 | 2648033 | C-T | HET | ||
| SBS | Chr6 | 3982552 | A-G | HET | ||
| SBS | Chr7 | 24134553 | G-T | HET | ||
| SBS | Chr8 | 14400145 | G-T | HET | ||
| SBS | Chr8 | 161509 | T-C | Homo | ||
| SBS | Chr8 | 21521246 | C-T | HET | ||
| SBS | Chr8 | 26041723 | C-T | HET | ||
| SBS | Chr8 | 26792846 | G-A | HET | ||
| SBS | Chr8 | 2852501 | G-T | HET | ||
| SBS | Chr8 | 3456889 | G-T | HET | ||
| SBS | Chr8 | 7641941 | C-T | HET | ||
| SBS | Chr8 | 7823297 | G-A | HET | ||
| SBS | Chr9 | 14946054 | C-A | HET | STOP_GAINED | LOC_Os09g25010 |
| SBS | Chr9 | 15554983 | T-C | HET | ||
| SBS | Chr9 | 17277792 | C-A | HET | ||
| SBS | Chr9 | 21539246 | C-A | HET | ||
| SBS | Chr9 | 6343340 | C-G | HET |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1929098 | 1929105 | 7 | |
| Deletion | Chr8 | 3117905 | 3117909 | 4 | |
| Deletion | Chr5 | 3235474 | 3235491 | 17 | |
| Deletion | Chr12 | 3378820 | 3378821 | 1 | |
| Deletion | Chr8 | 5845278 | 5845279 | 1 | |
| Deletion | Chr11 | 11710968 | 11710971 | 3 | LOC_Os11g20270 |
| Deletion | Chr10 | 12446001 | 12703000 | 256999 | 40 |
| Deletion | Chr10 | 12705001 | 12988000 | 282999 | 41 |
| Deletion | Chr4 | 13449001 | 13481000 | 31999 | 4 |
| Deletion | Chr7 | 14161198 | 14161202 | 4 | LOC_Os07g24910 |
| Deletion | Chr2 | 14627001 | 14639000 | 11999 | 2 |
| Deletion | Chr9 | 16771614 | 16771641 | 27 | |
| Deletion | Chr4 | 20520982 | 20520983 | 1 | |
| Deletion | Chr10 | 21565347 | 21565351 | 4 | |
| Deletion | Chr11 | 21709646 | 21709647 | 1 | |
| Deletion | Chr1 | 22709001 | 22725000 | 15999 | LOC_Os01g40250 |
| Deletion | Chr6 | 25262671 | 25262672 | 1 | |
| Deletion | Chr11 | 25824920 | 25824945 | 25 | |
| Deletion | Chr4 | 26198785 | 26198786 | 1 | |
| Deletion | Chr1 | 26992067 | 26992096 | 29 | LOC_Os01g47240 |
| Deletion | Chr11 | 27110835 | 27110837 | 2 | |
| Deletion | Chr6 | 28326001 | 28340000 | 13999 | 2 |
| Deletion | Chr2 | 31105937 | 31105950 | 13 | |
| Deletion | Chr2 | 34955412 | 34955416 | 4 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 26109535 | 26109536 | 2 |
No Inversion
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 4162371 | Chr1 | 19825688 | 2 |
| Translocation | Chr7 | 6388066 | Chr6 | 4161906 | 2 |
| Translocation | Chr8 | 15272138 | Chr3 | 1269592 | |
| Translocation | Chr9 | 19797986 | Chr6 | 4874166 | 2 |
| Translocation | Chr9 | 19798463 | Chr8 | 26888349 | 2 |
| Translocation | Chr9 | 19798465 | Chr8 | 26887481 | LOC_Os09g33559 |
| Translocation | Chr8 | 27804340 | Chr2 | 15724697 | 2 |
