Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2944-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2944-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 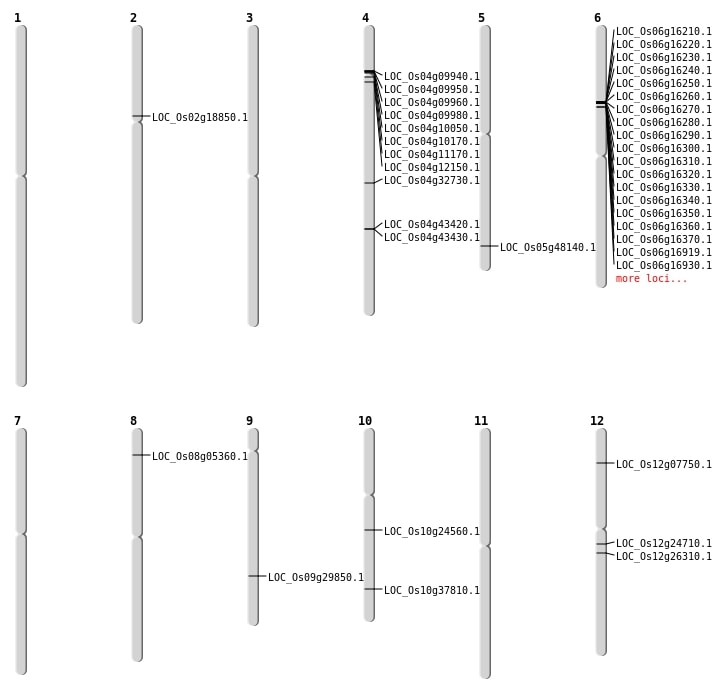
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 7916067 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 8104621 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 12608875 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24560 |
| SBS | Chr11 | 2728100 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 6353512 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 12664634 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 14169448 | C-T | HOMO | STOP_GAINED | LOC_Os12g24710 |
| SBS | Chr12 | 15356722 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g26310 |
| SBS | Chr12 | 1925315 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 3897800 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g07750 |
| SBS | Chr2 | 11000354 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g18850 |
| SBS | Chr2 | 14436034 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 190421 | C-T | HET | INTRON | |
| SBS | Chr2 | 20737756 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 25910004 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr2 | 29378067 | T-C | HOMO | INTRON | |
| SBS | Chr3 | 19192734 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 21832661 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 11333295 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 23590496 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 4805560 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 27597779 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g48140 |
| SBS | Chr5 | 4794471 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 6483681 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 10131089 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 21146951 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g36140 |
| SBS | Chr6 | 24124976 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 24180715 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 3902454 | A-G | HET | INTRON | |
| SBS | Chr6 | 3902465 | T-A | HET | INTRON | |
| SBS | Chr7 | 10633766 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 18022540 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 22710315 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 7516616 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 18804216 | C-T | HET | INTERGENIC |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1031110 | 1031111 | 1 | |
| Deletion | Chr8 | 1932671 | 1932672 | 1 | |
| Deletion | Chr1 | 2335893 | 2335894 | 1 | |
| Deletion | Chr12 | 4856637 | 4856638 | 1 | |
| Deletion | Chr9 | 4874595 | 4874597 | 2 | |
| Deletion | Chr4 | 5340001 | 5370000 | 29999 | 5 |
| Deletion | Chr9 | 5716599 | 5716609 | 10 | |
| Deletion | Chr1 | 5780664 | 5780666 | 2 | |
| Deletion | Chr11 | 6047809 | 6047810 | 1 | |
| Deletion | Chr4 | 6696262 | 6696263 | 1 | LOC_Os04g12150 |
| Deletion | Chr6 | 9222001 | 9348000 | 125999 | 17 |
| Deletion | Chr6 | 9547992 | 9548001 | 9 | |
| Deletion | Chr6 | 9601040 | 9601053 | 13 | |
| Deletion | Chr6 | 9771001 | 9786000 | 14999 | LOC_Os06g16919 |
| Deletion | Chr6 | 9802001 | 9956000 | 153999 | 24 |
| Deletion | Chr6 | 9987001 | 10095000 | 107999 | 16 |
| Deletion | Chr2 | 10851132 | 10851133 | 1 | |
| Deletion | Chr3 | 12267469 | 12267486 | 17 | |
| Deletion | Chr3 | 12340397 | 12340399 | 2 | |
| Deletion | Chr7 | 13176878 | 13176882 | 4 | |
| Deletion | Chr5 | 15242768 | 15242769 | 1 | |
| Deletion | Chr8 | 15544411 | 15544416 | 5 | |
| Deletion | Chr9 | 18156689 | 18156690 | 1 | LOC_Os09g29850 |
| Deletion | Chr8 | 18275080 | 18275083 | 3 | |
| Deletion | Chr10 | 18349758 | 18349759 | 1 | |
| Deletion | Chr10 | 20238493 | 20238496 | 3 | LOC_Os10g37810 |
| Deletion | Chr12 | 20389415 | 20389416 | 1 | |
| Deletion | Chr5 | 20603985 | 20604001 | 16 | |
| Deletion | Chr10 | 20809401 | 20809402 | 1 | |
| Deletion | Chr10 | 20984552 | 20984562 | 10 | |
| Deletion | Chr7 | 21545593 | 21545599 | 6 | |
| Deletion | Chr7 | 23109522 | 23109526 | 4 | |
| Deletion | Chr4 | 25681001 | 25706000 | 24999 | 2 |
| Deletion | Chr1 | 25816251 | 25816252 | 1 | |
| Deletion | Chr2 | 28355994 | 28355996 | 2 |
Insertions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 15865031 | 15865037 | 7 | |
| Insertion | Chr1 | 23475349 | 23475349 | 1 | |
| Insertion | Chr1 | 28275230 | 28275233 | 4 | |
| Insertion | Chr1 | 9031972 | 9031972 | 1 | |
| Insertion | Chr10 | 1301765 | 1301768 | 4 | |
| Insertion | Chr10 | 1416699 | 1416699 | 1 | |
| Insertion | Chr11 | 3211068 | 3211070 | 3 | |
| Insertion | Chr12 | 22456048 | 22456050 | 3 | |
| Insertion | Chr2 | 9205518 | 9205519 | 2 | |
| Insertion | Chr3 | 30018014 | 30018052 | 39 | |
| Insertion | Chr4 | 16556872 | 16556919 | 48 | |
| Insertion | Chr4 | 19731904 | 19731908 | 5 | LOC_Os04g32730 |
| Insertion | Chr4 | 23013617 | 23013617 | 1 | |
| Insertion | Chr4 | 5944437 | 5944452 | 16 | |
| Insertion | Chr5 | 12293134 | 12293134 | 1 | |
| Insertion | Chr6 | 9753636 | 9753636 | 1 | |
| Insertion | Chr7 | 24289012 | 24289013 | 2 | |
| Insertion | Chr7 | 8338880 | 8338881 | 2 | |
| Insertion | Chr8 | 2824391 | 2824391 | 1 | LOC_Os08g05360 |
| Insertion | Chr8 | 6625934 | 6625934 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 5338246 | 6073640 | LOC_Os04g11170 |
| Inversion | Chr4 | 26481779 | 26636639 | |
| Inversion | Chr4 | 26532270 | 26636634 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 1840332 | Chr3 | 30508219 | |
| Translocation | Chr8 | 5280456 | Chr4 | 5419644 | LOC_Os04g10050 |
| Translocation | Chr8 | 5280458 | Chr4 | 5419893 | LOC_Os04g10050 |
