Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2953-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2953-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 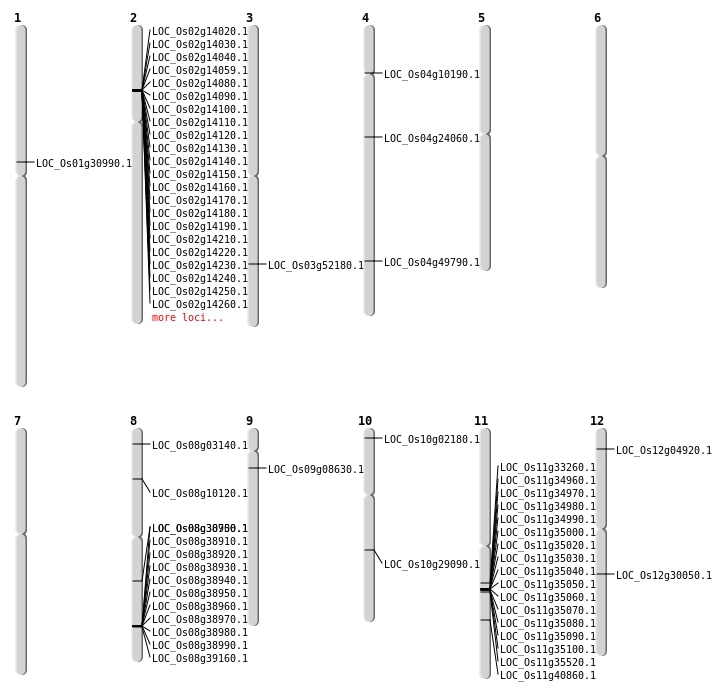
|
| Variant Information |
|---|
Single base substitutions: 26
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18744728 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 40969287 | G-T | HET | INTRON | |
| SBS | Chr1 | 4354068 | T-C | HET | INTRON | |
| SBS | Chr1 | 9889000 | C-A | HOMO | UTR_3_PRIME | |
| SBS | Chr10 | 22045490 | G-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 19662770 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g33260 |
| SBS | Chr11 | 19662771 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g33260 |
| SBS | Chr11 | 20473362 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 21578809 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 8360747 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 8717928 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 11216461 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 22168813 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 22383688 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g37050 |
| SBS | Chr2 | 30370446 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 32042403 | A-T | HOMO | INTRON | |
| SBS | Chr3 | 24152622 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 29958279 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g52180 |
| SBS | Chr4 | 13620641 | G-T | HET | INTRON | |
| SBS | Chr4 | 19682000 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr4 | 25905146 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 15173825 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 12494071 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 12971336 | G-C | HOMO | INTRON | |
| SBS | Chr9 | 8131888 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 8220863 | C-T | HOMO | INTERGENIC |
Deletions: 63
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 311478 | 311489 | 11 | |
| Deletion | Chr2 | 354839 | 354840 | 1 | |
| Deletion | Chr10 | 1255092 | 1255093 | 1 | |
| Deletion | Chr8 | 1441919 | 1441920 | 1 | LOC_Os08g03140 |
| Deletion | Chr12 | 2117055 | 2117056 | 1 | LOC_Os12g04920 |
| Deletion | Chr4 | 2857915 | 2857916 | 1 | |
| Deletion | Chr9 | 2858400 | 2858405 | 5 | |
| Deletion | Chr4 | 3248445 | 3248446 | 1 | |
| Deletion | Chr4 | 5510393 | 5510411 | 18 | LOC_Os04g10190 |
| Deletion | Chr10 | 5525714 | 5525719 | 5 | |
| Deletion | Chr10 | 5527876 | 5527877 | 1 | |
| Deletion | Chr8 | 5871009 | 5871012 | 3 | LOC_Os08g10120 |
| Deletion | Chr8 | 6378823 | 6378824 | 1 | |
| Deletion | Chr8 | 6395379 | 6395380 | 1 | |
| Deletion | Chr8 | 6973838 | 6973839 | 1 | |
| Deletion | Chr3 | 7071576 | 7071577 | 1 | |
| Deletion | Chr2 | 7642001 | 7829000 | 186999 | 23 |
| Deletion | Chr9 | 7860427 | 7860466 | 39 | |
| Deletion | Chr8 | 8439030 | 8439031 | 1 | |
| Deletion | Chr8 | 8740457 | 8740460 | 3 | |
| Deletion | Chr6 | 9104038 | 9104040 | 2 | |
| Deletion | Chr4 | 9253948 | 9253949 | 1 | |
| Deletion | Chr11 | 9268246 | 9268250 | 4 | |
| Deletion | Chr4 | 10481899 | 10481900 | 1 | |
| Deletion | Chr7 | 12249760 | 12249761 | 1 | |
| Deletion | Chr12 | 12333270 | 12333273 | 3 | |
| Deletion | Chr1 | 13632947 | 13632948 | 1 | |
| Deletion | Chr4 | 13744715 | 13744727 | 12 | LOC_Os04g24060 |
| Deletion | Chr1 | 14291369 | 14291379 | 10 | |
| Deletion | Chr11 | 14447008 | 14447009 | 1 | |
| Deletion | Chr2 | 14519494 | 14519495 | 1 | |
| Deletion | Chr12 | 14852262 | 14852281 | 19 | |
| Deletion | Chr8 | 15302134 | 15302138 | 4 | |
| Deletion | Chr12 | 15598351 | 15598365 | 14 | |
| Deletion | Chr2 | 15666557 | 15666558 | 1 | |
| Deletion | Chr3 | 15859712 | 15859714 | 2 | |
| Deletion | Chr9 | 16373266 | 16373269 | 3 | |
| Deletion | Chr11 | 16724643 | 16724644 | 1 | |
| Deletion | Chr1 | 16939636 | 16939645 | 9 | LOC_Os01g30990 |
| Deletion | Chr8 | 17046460 | 17046461 | 1 | |
| Deletion | Chr12 | 17996187 | 17996188 | 1 | LOC_Os12g30050 |
| Deletion | Chr8 | 18970871 | 18970895 | 24 | LOC_Os08g30750 |
| Deletion | Chr6 | 19641970 | 19641971 | 1 | |
| Deletion | Chr1 | 19809425 | 19809447 | 22 | |
| Deletion | Chr12 | 20240099 | 20240100 | 1 | |
| Deletion | Chr5 | 20358272 | 20358273 | 1 | |
| Deletion | Chr11 | 20479001 | 20582000 | 102999 | 15 |
| Deletion | Chr9 | 21444277 | 21444278 | 1 | |
| Deletion | Chr3 | 21462544 | 21462545 | 1 | |
| Deletion | Chr12 | 22111606 | 22111609 | 3 | |
| Deletion | Chr11 | 22525106 | 22525107 | 1 | |
| Deletion | Chr2 | 22614136 | 22614148 | 12 | |
| Deletion | Chr12 | 22805945 | 22805962 | 17 | |
| Deletion | Chr5 | 24373217 | 24373239 | 22 | |
| Deletion | Chr8 | 24581001 | 24647000 | 65999 | 11 |
| Deletion | Chr4 | 26413536 | 26413537 | 1 | |
| Deletion | Chr8 | 27201448 | 27201456 | 8 | |
| Deletion | Chr11 | 27293985 | 27293986 | 1 | |
| Deletion | Chr1 | 27436600 | 27436601 | 1 | |
| Deletion | Chr4 | 28947850 | 28947856 | 6 | |
| Deletion | Chr5 | 29849533 | 29849887 | 354 | |
| Deletion | Chr3 | 34749902 | 34749904 | 2 | |
| Deletion | Chr1 | 43264101 | 43264108 | 7 |
Insertions: 48
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11390065 | 11390065 | 1 | |
| Insertion | Chr1 | 15683389 | 15683486 | 98 | |
| Insertion | Chr1 | 35974726 | 35974730 | 5 | |
| Insertion | Chr10 | 15056605 | 15056605 | 1 | |
| Insertion | Chr10 | 15151619 | 15151641 | 23 | LOC_Os10g29090 |
| Insertion | Chr10 | 3209977 | 3209977 | 1 | |
| Insertion | Chr10 | 749754 | 749754 | 1 | LOC_Os10g02180 |
| Insertion | Chr10 | 7669829 | 7669875 | 47 | |
| Insertion | Chr10 | 8724646 | 8724646 | 1 | |
| Insertion | Chr11 | 21228297 | 21228297 | 1 | |
| Insertion | Chr12 | 11840819 | 11840864 | 46 | |
| Insertion | Chr12 | 14426298 | 14426299 | 2 | |
| Insertion | Chr12 | 16231854 | 16231854 | 1 | |
| Insertion | Chr12 | 23764592 | 23764684 | 93 | |
| Insertion | Chr12 | 4584502 | 4584507 | 6 | |
| Insertion | Chr12 | 5156788 | 5156789 | 2 | |
| Insertion | Chr2 | 11908506 | 11908506 | 1 | |
| Insertion | Chr2 | 13728134 | 13728134 | 1 | |
| Insertion | Chr3 | 13532182 | 13532182 | 1 | |
| Insertion | Chr3 | 24089602 | 24089603 | 2 | |
| Insertion | Chr4 | 24450085 | 24450085 | 1 | |
| Insertion | Chr4 | 26164892 | 26164893 | 2 | |
| Insertion | Chr4 | 3332318 | 3332319 | 2 | |
| Insertion | Chr4 | 33350031 | 33350031 | 1 | |
| Insertion | Chr4 | 7551233 | 7551335 | 103 | |
| Insertion | Chr4 | 8027137 | 8027137 | 1 | |
| Insertion | Chr4 | 8576183 | 8576185 | 3 | |
| Insertion | Chr4 | 8745059 | 8745064 | 6 | |
| Insertion | Chr4 | 9840807 | 9840807 | 1 | |
| Insertion | Chr5 | 11556600 | 11556600 | 1 | |
| Insertion | Chr5 | 27014414 | 27014414 | 1 | |
| Insertion | Chr5 | 29411363 | 29411404 | 42 | |
| Insertion | Chr5 | 29536988 | 29536990 | 3 | |
| Insertion | Chr5 | 4919066 | 4919066 | 1 | |
| Insertion | Chr5 | 9930890 | 9930891 | 2 | |
| Insertion | Chr6 | 22187968 | 22187968 | 1 | |
| Insertion | Chr6 | 22679281 | 22679373 | 93 | |
| Insertion | Chr7 | 13577283 | 13577283 | 1 | |
| Insertion | Chr7 | 27613461 | 27613462 | 2 | |
| Insertion | Chr8 | 16867262 | 16867262 | 1 | |
| Insertion | Chr8 | 5509500 | 5509500 | 1 | |
| Insertion | Chr8 | 6459106 | 6459195 | 90 | |
| Insertion | Chr8 | 6945198 | 6945198 | 1 | |
| Insertion | Chr8 | 6973635 | 6973636 | 2 | |
| Insertion | Chr9 | 12053404 | 12053404 | 1 | |
| Insertion | Chr9 | 12953844 | 12953848 | 5 | |
| Insertion | Chr9 | 4514786 | 4514869 | 84 | LOC_Os09g08630 |
| Insertion | Chr9 | 9845550 | 9845550 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 19147967 | 19888647 | |
| Inversion | Chr11 | 20481894 | 20813279 |
Translocations: 19
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 4009896 | Chr6 | 23504877 | |
| Translocation | Chr9 | 5190823 | Chr6 | 8717945 | |
| Translocation | Chr10 | 7047390 | Chr1 | 36582409 | |
| Translocation | Chr10 | 11149694 | Chr9 | 9850710 | |
| Translocation | Chr12 | 11421780 | Chr1 | 42198377 | |
| Translocation | Chr12 | 11422019 | Chr1 | 42198373 | |
| Translocation | Chr11 | 12004248 | Chr10 | 2306195 | |
| Translocation | Chr9 | 13620982 | Chr4 | 7872859 | |
| Translocation | Chr8 | 15935206 | Chr7 | 6242593 | |
| Translocation | Chr11 | 18148243 | Chr6 | 6320469 | |
| Translocation | Chr11 | 18250795 | Chr1 | 26330533 | |
| Translocation | Chr7 | 19888661 | Chr6 | 1555753 | |
| Translocation | Chr7 | 19888811 | Chr6 | 1555752 | |
| Translocation | Chr11 | 20281824 | Chr2 | 13966343 | LOC_Os02g24100 |
| Translocation | Chr7 | 20707332 | Chr6 | 518657 | |
| Translocation | Chr6 | 23392560 | Chr2 | 20261349 | |
| Translocation | Chr11 | 24445386 | Chr4 | 29686988 | 2 |
| Translocation | Chr8 | 24645782 | Chr7 | 19888832 | |
| Translocation | Chr3 | 32039583 | Chr2 | 5535368 |
