Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3178-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3178-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 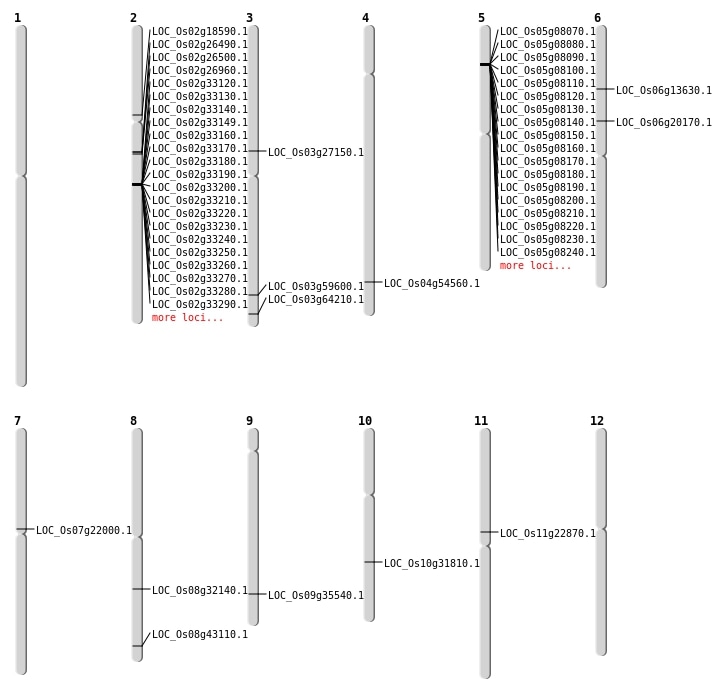
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22760753 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 7144034 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 17198150 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 11189535 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 13153141 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22870 |
| SBS | Chr11 | 23260250 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 9086054 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 9522640 | C-A | HET | INTRON | |
| SBS | Chr11 | 9522641 | C-A | HET | INTRON | |
| SBS | Chr12 | 21150401 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 25773386 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 9000569 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 10838677 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g18590 |
| SBS | Chr2 | 32466739 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g53060 |
| SBS | Chr3 | 15545178 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g27150 |
| SBS | Chr3 | 33937927 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g59600 |
| SBS | Chr3 | 34742395 | G-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 2339457 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 24991903 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 11300589 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 15983880 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 8961962 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 7548773 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g13630 |
| SBS | Chr7 | 17676075 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 22680040 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 10421090 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 19943769 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g32140 |
| SBS | Chr8 | 19943770 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g32140 |
| SBS | Chr8 | 19987271 | T-A | HOMO | INTRON | |
| SBS | Chr8 | 23722915 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 27735664 | T-A | HET | INTRON | |
| SBS | Chr9 | 12928524 | G-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 20423278 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g35540 |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 39071 | 39090 | 19 | |
| Deletion | Chr6 | 4299116 | 4299117 | 1 | |
| Deletion | Chr5 | 4385001 | 4399000 | 13999 | 3 |
| Deletion | Chr5 | 4402001 | 4504000 | 101999 | 17 |
| Deletion | Chr11 | 4540580 | 4540582 | 2 | |
| Deletion | Chr3 | 4631015 | 4631023 | 8 | |
| Deletion | Chr2 | 10851132 | 10851133 | 1 | |
| Deletion | Chr7 | 12305397 | 12305474 | 77 | LOC_Os07g22000 |
| Deletion | Chr9 | 12954529 | 12954533 | 4 | |
| Deletion | Chr2 | 13318712 | 13318713 | 1 | |
| Deletion | Chr12 | 14840140 | 14840144 | 4 | |
| Deletion | Chr2 | 15551001 | 15564000 | 12999 | 3 |
| Deletion | Chr1 | 16788197 | 16788198 | 1 | |
| Deletion | Chr8 | 17884276 | 17884280 | 4 | |
| Deletion | Chr2 | 18426070 | 18426071 | 1 | |
| Deletion | Chr7 | 18936645 | 18936711 | 66 | |
| Deletion | Chr11 | 18955921 | 18955923 | 2 | |
| Deletion | Chr2 | 19683001 | 20207000 | 523999 | 77 |
| Deletion | Chr2 | 20213001 | 20405000 | 191999 | 17 |
| Deletion | Chr2 | 20407001 | 20518000 | 110999 | 17 |
| Deletion | Chr2 | 20529001 | 20561000 | 31999 | 6 |
| Deletion | Chr2 | 24021363 | 24021365 | 2 | |
| Deletion | Chr8 | 24988095 | 24988098 | 3 | |
| Deletion | Chr6 | 27791668 | 27791669 | 1 | |
| Deletion | Chr3 | 35577562 | 35577567 | 5 | |
| Deletion | Chr3 | 36283643 | 36283647 | 4 | LOC_Os03g64210 |
| Deletion | Chr1 | 36817241 | 36817242 | 1 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 2937965 | 2937965 | 1 | |
| Insertion | Chr4 | 10337162 | 10337162 | 1 | |
| Insertion | Chr4 | 25956503 | 25956503 | 1 | |
| Insertion | Chr5 | 21189414 | 21189414 | 1 | |
| Insertion | Chr8 | 24873234 | 24873234 | 1 | |
| Insertion | Chr8 | 27255845 | 27255845 | 1 | LOC_Os08g43110 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 4379038 | 5033664 | |
| Inversion | Chr5 | 5033674 | 5404076 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 1493893 | Chr5 | 10967042 | LOC_Os05g18890 |
| Translocation | Chr10 | 3169145 | Chr8 | 16573963 | |
| Translocation | Chr11 | 5348583 | Chr10 | 19020825 | |
| Translocation | Chr12 | 7267589 | Chr6 | 20611018 | |
| Translocation | Chr6 | 11582952 | Chr2 | 15565012 | LOC_Os02g26500 |
| Translocation | Chr10 | 16676156 | Chr6 | 11570672 | 2 |
| Translocation | Chr8 | 17951052 | Chr7 | 27365777 | |
| Translocation | Chr5 | 19226218 | Chr4 | 32449936 | LOC_Os04g54560 |
| Translocation | Chr2 | 20743116 | Chr1 | 2919224 | |
| Translocation | Chr4 | 28579316 | Chr1 | 18061446 | |
| Translocation | Chr4 | 28585784 | Chr1 | 18061442 |
