Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3185-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3185-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 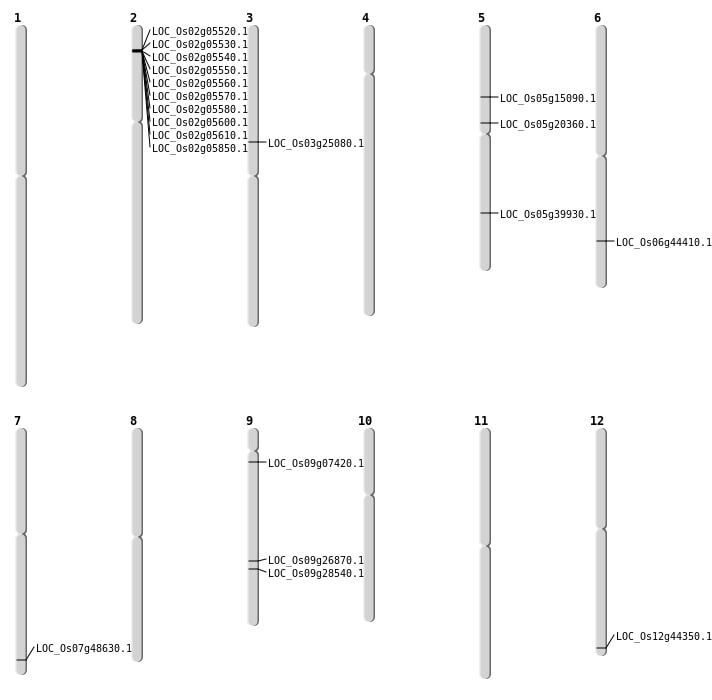
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 28566540 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 5486005 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 22389569 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 5813184 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 5813185 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 23541306 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 23686766 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 26505355 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 26505357 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 3776312 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 4375134 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 28210316 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 32890447 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 34059676 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 7825985 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 14324969 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g25080 |
| SBS | Chr3 | 19846039 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 6549370 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 10792584 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 12122074 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 15996966 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 24062778 | G-T | HET | INTRON | |
| SBS | Chr4 | 26357228 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 35274577 | T-C | HET | INTRON | |
| SBS | Chr4 | 35274581 | G-A | HET | INTRON | |
| SBS | Chr4 | 5376828 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 11937331 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g20360 |
| SBS | Chr5 | 17243544 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 27051730 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 8567704 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g15090 |
| SBS | Chr6 | 26810844 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g44410 |
| SBS | Chr7 | 14026266 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 15957797 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 832327 | C-T | HET | INTRON | |
| SBS | Chr8 | 11012988 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 251515 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 4411673 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 16329569 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g26870 |
| SBS | Chr9 | 18624899 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 3698061 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g07420 |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 56201 | 56202 | 1 | |
| Deletion | Chr10 | 166206 | 166210 | 4 | |
| Deletion | Chr9 | 206099 | 206102 | 3 | |
| Deletion | Chr7 | 215989 | 215991 | 2 | |
| Deletion | Chr8 | 963282 | 963303 | 21 | |
| Deletion | Chr6 | 1575790 | 1575795 | 5 | |
| Deletion | Chr4 | 1954027 | 1954041 | 14 | |
| Deletion | Chr2 | 2664001 | 2709000 | 44999 | 8 |
| Deletion | Chr2 | 2713001 | 2733000 | 19999 | 2 |
| Deletion | Chr3 | 3277732 | 3277735 | 3 | |
| Deletion | Chr4 | 3522088 | 3522158 | 70 | |
| Deletion | Chr7 | 4565010 | 4565013 | 3 | |
| Deletion | Chr7 | 4622879 | 4622880 | 1 | |
| Deletion | Chr6 | 5162589 | 5162591 | 2 | |
| Deletion | Chr10 | 8477114 | 8477127 | 13 | |
| Deletion | Chr9 | 8548251 | 8548258 | 7 | |
| Deletion | Chr6 | 12216078 | 12216121 | 43 | |
| Deletion | Chr9 | 15181387 | 15181389 | 2 | |
| Deletion | Chr5 | 15968074 | 15968076 | 2 | |
| Deletion | Chr12 | 16018037 | 16018042 | 5 | |
| Deletion | Chr10 | 16172641 | 16172651 | 10 | |
| Deletion | Chr9 | 16214876 | 16214878 | 2 | |
| Deletion | Chr9 | 17357202 | 17357212 | 10 | LOC_Os09g28540 |
| Deletion | Chr10 | 17505608 | 17505622 | 14 | |
| Deletion | Chr3 | 18026433 | 18026434 | 1 | |
| Deletion | Chr1 | 18283076 | 18283077 | 1 | |
| Deletion | Chr4 | 18370353 | 18370357 | 4 | |
| Deletion | Chr9 | 22881845 | 22881848 | 3 | |
| Deletion | Chr5 | 23464767 | 23464773 | 6 | LOC_Os05g39930 |
| Deletion | Chr12 | 27493068 | 27493073 | 5 | LOC_Os12g44350 |
| Deletion | Chr1 | 33990857 | 33990858 | 1 | |
| Deletion | Chr1 | 34058206 | 34058216 | 10 | |
| Deletion | Chr4 | 34108120 | 34108123 | 3 |
Insertions: 5
No Inversion
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 3091495 | Chr1 | 41813589 | |
| Translocation | Chr7 | 11086758 | Chr4 | 32086848 | |
| Translocation | Chr11 | 17795284 | Chr7 | 29118626 | LOC_Os07g48630 |
| Translocation | Chr11 | 19757340 | Chr1 | 33088407 |
