Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN319-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN319-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 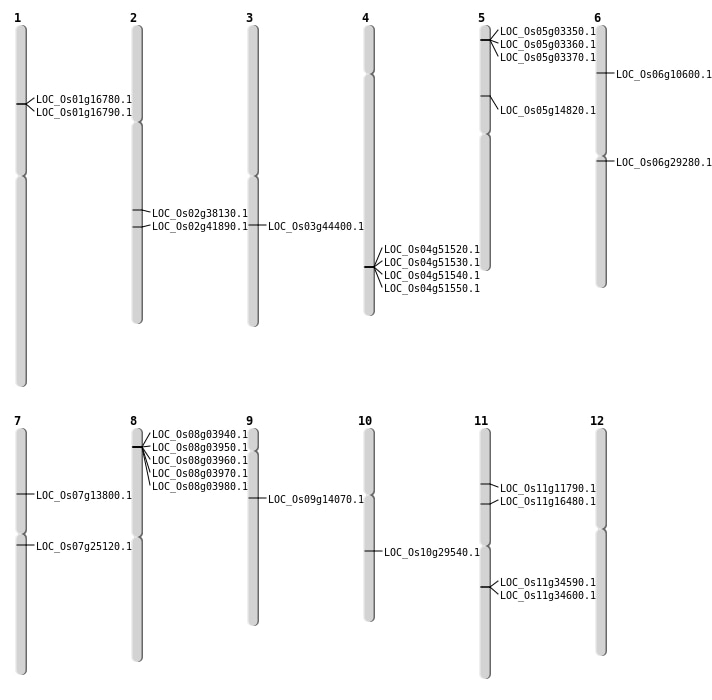
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17894348 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 34059791 | G-T | HET | INTRON | |
| SBS | Chr1 | 6997824 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 12394707 | C-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 13190333 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 15348121 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g29540 |
| SBS | Chr11 | 11500174 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 1023176 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 22476872 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 10764178 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 10764179 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 25176555 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g41890 |
| SBS | Chr2 | 26024606 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 24981623 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g44400 |
| SBS | Chr4 | 13761306 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 149304 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 33358834 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 15547316 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 6684122 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 16716694 | G-A | HET | STOP_GAINED | LOC_Os06g29280 |
| SBS | Chr6 | 5257785 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 5501385 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g10600 |
| SBS | Chr7 | 14348653 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g25120 |
| SBS | Chr7 | 28751629 | A-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 7909280 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g13800 |
| SBS | Chr7 | 8716231 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 9325050 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr8 | 19209840 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 21310522 | G-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 21717319 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 8330241 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g14070 |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 674101 | 674103 | 2 | |
| Deletion | Chr5 | 1374001 | 1392000 | 18000 | 3 |
| Deletion | Chr8 | 1876001 | 1914000 | 38000 | 5 |
| Deletion | Chr12 | 2190933 | 2190946 | 13 | |
| Deletion | Chr9 | 2546177 | 2546178 | 1 | |
| Deletion | Chr1 | 3571431 | 3571436 | 5 | |
| Deletion | Chr11 | 6543412 | 6543440 | 28 | LOC_Os11g11790 |
| Deletion | Chr4 | 6667085 | 6667086 | 1 | |
| Deletion | Chr5 | 6792262 | 6792279 | 17 | |
| Deletion | Chr11 | 9116818 | 9116820 | 2 | LOC_Os11g16480 |
| Deletion | Chr1 | 9531001 | 9544000 | 13000 | 2 |
| Deletion | Chr7 | 9771908 | 9771916 | 8 | |
| Deletion | Chr12 | 11128174 | 11128205 | 31 | |
| Deletion | Chr2 | 13605031 | 13605037 | 6 | |
| Deletion | Chr7 | 18474153 | 18474161 | 8 | |
| Deletion | Chr8 | 18733503 | 18733504 | 1 | |
| Deletion | Chr7 | 20202278 | 20202295 | 17 | |
| Deletion | Chr11 | 20242001 | 20255000 | 13000 | 2 |
| Deletion | Chr4 | 20679554 | 20679560 | 6 | |
| Deletion | Chr6 | 20933151 | 20933174 | 23 | |
| Deletion | Chr8 | 21109672 | 21109685 | 13 | |
| Deletion | Chr7 | 21538389 | 21538390 | 1 | |
| Deletion | Chr2 | 23062974 | 23062989 | 15 | LOC_Os02g38130 |
| Deletion | Chr1 | 26588646 | 26588647 | 1 | |
| Deletion | Chr4 | 30523001 | 30538000 | 15000 | 4 |
| Deletion | Chr2 | 30568205 | 30568206 | 1 |
Insertions: 5
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 8428042 | Chr1 | 23528673 | LOC_Os05g14820 |
| Translocation | Chr7 | 15550475 | Chr5 | 5643362 |
