Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3243-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3243-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 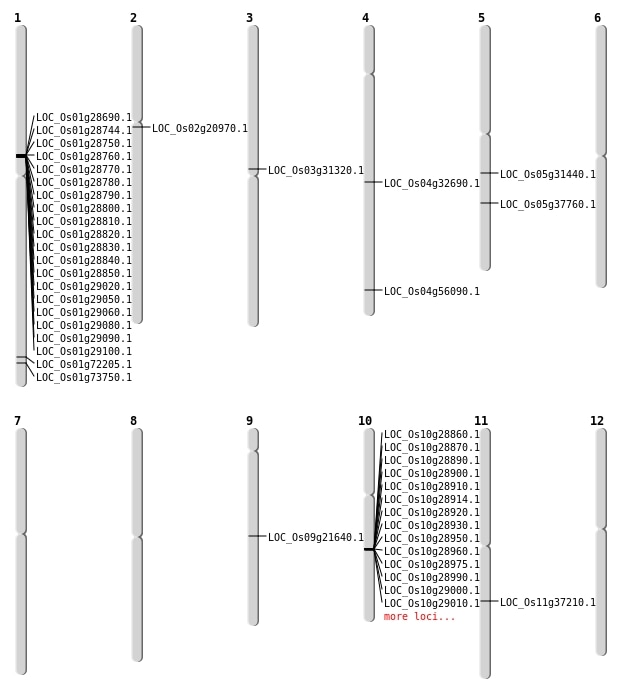
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 4160156 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 41867951 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g72205 |
| SBS | Chr1 | 42721542 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g73750 |
| SBS | Chr1 | 4276503 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 4276504 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 19721820 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 20167057 | G-T | HET | INTRON | |
| SBS | Chr11 | 16471874 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 21987460 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g37210 |
| SBS | Chr11 | 3289392 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 4739444 | G-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 13318326 | C-T | HET | INTRON | |
| SBS | Chr12 | 16938955 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 19117635 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 19117638 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 19117642 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 2157812 | G-A | HET | INTRON | |
| SBS | Chr12 | 6952552 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 24881914 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 12791573 | T-C | HET | INTRON | |
| SBS | Chr3 | 1305381 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 20579396 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 35737691 | G-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr4 | 10648883 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 19709860 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g32690 |
| SBS | Chr4 | 19709861 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g32690 |
| SBS | Chr5 | 18622169 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 20959559 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 22104976 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g37760 |
| SBS | Chr5 | 8635485 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 1879603 | T-C | HET | INTRON | |
| SBS | Chr6 | 20250274 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 6022986 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 21303461 | C-T | HET | INTRON | |
| SBS | Chr7 | 25401629 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 25524878 | G-T | HET | START_GAINED | |
| SBS | Chr8 | 12112316 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 22344046 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 3926933 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 19797669 | C-T | HET | INTRON |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 716390 | 716398 | 8 | |
| Deletion | Chr11 | 1575536 | 1575537 | 1 | |
| Deletion | Chr8 | 1594378 | 1594379 | 1 | |
| Deletion | Chr7 | 3411011 | 3411016 | 5 | |
| Deletion | Chr5 | 5504336 | 5504339 | 3 | |
| Deletion | Chr4 | 9420650 | 9420662 | 12 | |
| Deletion | Chr12 | 11917764 | 11917782 | 18 | |
| Deletion | Chr2 | 12404894 | 12404895 | 1 | LOC_Os02g20970 |
| Deletion | Chr9 | 13105235 | 13105236 | 1 | LOC_Os09g21640 |
| Deletion | Chr1 | 14028969 | 14028970 | 1 | |
| Deletion | Chr10 | 15037001 | 15313000 | 275999 | 57 |
| Deletion | Chr10 | 15179717 | 15179743 | 26 | |
| Deletion | Chr1 | 16093001 | 16157000 | 63999 | 13 |
| Deletion | Chr1 | 16244001 | 16253000 | 8999 | LOC_Os01g29050 |
| Deletion | Chr1 | 16261001 | 16269000 | 7999 | 2 |
| Deletion | Chr1 | 16279001 | 16293000 | 13999 | 3 |
| Deletion | Chr5 | 16343721 | 16343747 | 26 | |
| Deletion | Chr8 | 16384425 | 16384481 | 56 | |
| Deletion | Chr4 | 16408632 | 16408693 | 61 | |
| Deletion | Chr11 | 17306095 | 17306097 | 2 | |
| Deletion | Chr3 | 17851451 | 17851452 | 1 | LOC_Os03g31320 |
| Deletion | Chr5 | 18284474 | 18284479 | 5 | LOC_Os05g31440 |
| Deletion | Chr8 | 18469492 | 18469505 | 13 | |
| Deletion | Chr8 | 21109551 | 21109553 | 2 | |
| Deletion | Chr9 | 21123410 | 21123460 | 50 | |
| Deletion | Chr2 | 21459149 | 21459150 | 1 | |
| Deletion | Chr12 | 22226752 | 22226753 | 1 | |
| Deletion | Chr11 | 27312050 | 27312053 | 3 | |
| Deletion | Chr11 | 27676960 | 27676986 | 26 | |
| Deletion | Chr6 | 28193488 | 28193489 | 1 | |
| Deletion | Chr4 | 31708736 | 31708737 | 1 | |
| Deletion | Chr4 | 33408029 | 33408046 | 17 | LOC_Os04g56090 |
| Deletion | Chr2 | 34039814 | 34039816 | 2 | |
| Deletion | Chr1 | 36027282 | 36027292 | 10 |
Insertions: 12
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 3079308 | 3174698 | |
| Inversion | Chr4 | 11474272 | 11474560 | |
| Inversion | Chr1 | 16062208 | 16156957 | LOC_Os01g28690 |
| Inversion | Chr1 | 16062209 | 16244372 | LOC_Os01g28690 |
| Inversion | Chr1 | 16156983 | 16292147 | LOC_Os01g28850 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 8867765 | Chr1 | 16062213 | LOC_Os01g28690 |
| Translocation | Chr10 | 15297278 | Chr4 | 4108314 | |
| Translocation | Chr10 | 18634800 | Chr8 | 27836248 | |
| Translocation | Chr11 | 27253533 | Chr7 | 22208108 |
