Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3269-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3269-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 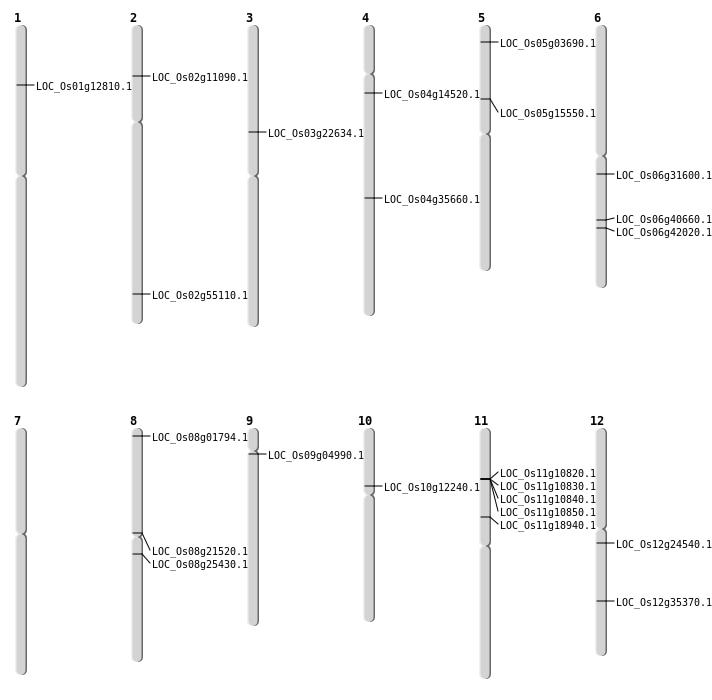
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19636654 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 39221861 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 20701862 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 3170971 | C-A | HOMO | INTRON | |
| SBS | Chr11 | 10201811 | C-T | HET | INTRON | |
| SBS | Chr11 | 10201812 | A-T | HET | INTRON | |
| SBS | Chr11 | 14900939 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 16467653 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 25157396 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 26386269 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 9620207 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 21507933 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g35370 |
| SBS | Chr12 | 5524829 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 33752586 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g55110 |
| SBS | Chr3 | 24211145 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 1663557 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 17597352 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 2623977 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 29884868 | A-G | HOMO | START_GAINED | |
| SBS | Chr4 | 5500839 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 8131993 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g14520 |
| SBS | Chr5 | 1605154 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 1605156 | A-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g03690 |
| SBS | Chr5 | 8152718 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 8799450 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g15550 |
| SBS | Chr6 | 26068223 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 6969837 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 6801187 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 10878261 | T-C | HET | INTRON | |
| SBS | Chr8 | 12822436 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g21520 |
| SBS | Chr8 | 15479022 | A-T | HET | SPLICE_SITE_ACCEPTOR | LOC_Os08g25430 |
| SBS | Chr8 | 1581310 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr8 | 1581312 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr8 | 478398 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g01794 |
| SBS | Chr9 | 13660829 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 17959141 | C-T | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 828940 | 828944 | 4 | |
| Deletion | Chr8 | 1777864 | 1777865 | 1 | |
| Deletion | Chr9 | 2687807 | 2687809 | 2 | LOC_Os09g04990 |
| Deletion | Chr8 | 3911768 | 3911773 | 5 | |
| Deletion | Chr4 | 5039234 | 5039236 | 2 | |
| Deletion | Chr11 | 5969001 | 5980000 | 10999 | 4 |
| Deletion | Chr9 | 6891538 | 6891547 | 9 | |
| Deletion | Chr1 | 7093071 | 7093076 | 5 | LOC_Os01g12810 |
| Deletion | Chr7 | 9766977 | 9766982 | 5 | |
| Deletion | Chr7 | 10446430 | 10446431 | 1 | |
| Deletion | Chr6 | 11800947 | 11800952 | 5 | |
| Deletion | Chr3 | 13047180 | 13047185 | 5 | |
| Deletion | Chr3 | 13064040 | 13064044 | 4 | LOC_Os03g22634 |
| Deletion | Chr7 | 13523651 | 13523653 | 2 | |
| Deletion | Chr3 | 13655699 | 13655700 | 1 | |
| Deletion | Chr12 | 14018603 | 14018614 | 11 | LOC_Os12g24540 |
| Deletion | Chr4 | 19544456 | 19544464 | 8 | |
| Deletion | Chr4 | 21734441 | 21734446 | 5 | LOC_Os04g35660 |
| Deletion | Chr10 | 22025980 | 22025989 | 9 | |
| Deletion | Chr8 | 22593874 | 22593907 | 33 | |
| Deletion | Chr6 | 24242466 | 24242472 | 6 | LOC_Os06g40660 |
| Deletion | Chr3 | 24765332 | 24765334 | 2 | |
| Deletion | Chr6 | 25231425 | 25231426 | 1 | LOC_Os06g42020 |
| Deletion | Chr2 | 25334190 | 25334202 | 12 | |
| Deletion | Chr8 | 25691846 | 25691855 | 9 | |
| Deletion | Chr7 | 25780636 | 25780644 | 8 | |
| Deletion | Chr3 | 31579868 | 31579870 | 2 | |
| Deletion | Chr1 | 31639651 | 31639677 | 26 | |
| Deletion | Chr1 | 43187790 | 43187800 | 10 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr5 | 25089464 | 25089473 | 10 | |
| Insertion | Chr7 | 24240105 | 24240183 | 79 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 5966126 | 5969437 | LOC_Os11g10820 |
| Inversion | Chr11 | 9840757 | 9912542 | |
| Inversion | Chr11 | 9912528 | 10442374 | |
| Inversion | Chr11 | 10201001 | 10219655 | |
| Inversion | Chr11 | 10219662 | 10442830 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 1302556 | Chr2 | 15512942 | |
| Translocation | Chr11 | 6602817 | Chr2 | 4170016 | |
| Translocation | Chr10 | 6817373 | Chr6 | 18374054 | 2 |
| Translocation | Chr12 | 9418700 | Chr1 | 818514 | |
| Translocation | Chr11 | 10201011 | Chr5 | 22349550 | |
| Translocation | Chr11 | 10201166 | Chr5 | 22349551 | |
| Translocation | Chr11 | 10874570 | Chr2 | 5944591 | LOC_Os02g11090 |
| Translocation | Chr11 | 11900466 | Chr9 | 13703331 | |
| Translocation | Chr9 | 15256577 | Chr8 | 12780720 | |
| Translocation | Chr6 | 24570067 | Chr2 | 18235196 |
