Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3310-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3310-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 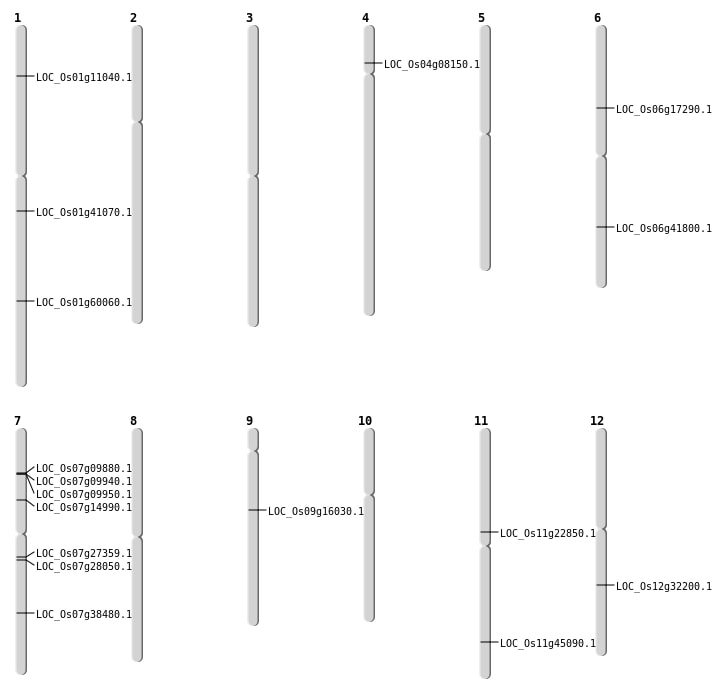
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23259995 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g41070 |
| SBS | Chr1 | 34743488 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g60060 |
| SBS | Chr1 | 5896544 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g11040 |
| SBS | Chr10 | 19418877 | C-T | Homo | ||
| SBS | Chr10 | 5193610 | T-G | HET | ||
| SBS | Chr10 | 7054395 | C-G | HET | ||
| SBS | Chr10 | 9578608 | G-T | HET | ||
| SBS | Chr11 | 13860394 | C-T | HET | ||
| SBS | Chr11 | 21935866 | G-A | HET | ||
| SBS | Chr11 | 22380124 | A-G | HET | ||
| SBS | Chr11 | 6401095 | C-A | HET | ||
| SBS | Chr12 | 19434984 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g32200 |
| SBS | Chr12 | 24071653 | A-T | HET | ||
| SBS | Chr2 | 12414148 | T-C | HET | ||
| SBS | Chr2 | 19079096 | G-A | HET | ||
| SBS | Chr2 | 19079097 | T-A | HET | ||
| SBS | Chr2 | 30491525 | G-A | HET | ||
| SBS | Chr2 | 32922750 | G-A | HET | ||
| SBS | Chr2 | 34643205 | A-T | HET | ||
| SBS | Chr2 | 3819737 | A-C | HET | ||
| SBS | Chr3 | 12171836 | A-T | HET | ||
| SBS | Chr3 | 13104423 | G-A | HET | ||
| SBS | Chr3 | 2684805 | C-T | HET | ||
| SBS | Chr3 | 6998592 | G-C | HET | ||
| SBS | Chr4 | 1594507 | T-C | HET | ||
| SBS | Chr4 | 19179804 | T-C | Homo | ||
| SBS | Chr4 | 23297364 | A-T | HET | ||
| SBS | Chr4 | 24719524 | C-A | HET | ||
| SBS | Chr4 | 35100663 | A-G | Homo | ||
| SBS | Chr4 | 4340146 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g08150 |
| SBS | Chr5 | 12331860 | T-C | HET | ||
| SBS | Chr5 | 13003698 | T-G | HET | ||
| SBS | Chr6 | 10015642 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g17290 |
| SBS | Chr6 | 19665781 | G-A | HET | ||
| SBS | Chr6 | 25069517 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g41800 |
| SBS | Chr7 | 14679127 | G-A | HET | ||
| SBS | Chr7 | 15925622 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g27359 |
| SBS | Chr7 | 16355650 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g28050 |
| SBS | Chr7 | 16615543 | G-A | HET | ||
| SBS | Chr7 | 20889535 | G-T | Homo | ||
| SBS | Chr7 | 27845483 | T-A | HET | ||
| SBS | Chr7 | 2980084 | C-T | HET | ||
| SBS | Chr7 | 5253902 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g09880 |
| SBS | Chr7 | 8595415 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g14990 |
| SBS | Chr8 | 11835499 | C-G | Homo | ||
| SBS | Chr8 | 18230221 | G-A | HET | ||
| SBS | Chr8 | 4124019 | A-T | HET | ||
| SBS | Chr9 | 11583398 | C-T | HET | ||
| SBS | Chr9 | 12043411 | A-G | HET | ||
| SBS | Chr9 | 9791393 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g16030 |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 174891 | 174892 | 1 | |
| Deletion | Chr4 | 351665 | 351667 | 2 | |
| Deletion | Chr12 | 2266988 | 2266995 | 7 | |
| Deletion | Chr6 | 3799462 | 3799472 | 10 | |
| Deletion | Chr7 | 5291001 | 5304000 | 12999 | 2 |
| Deletion | Chr7 | 5439624 | 5439625 | 1 | |
| Deletion | Chr1 | 5480764 | 5480766 | 2 | |
| Deletion | Chr7 | 14146831 | 14146832 | 1 | |
| Deletion | Chr12 | 14727440 | 14727443 | 3 | |
| Deletion | Chr9 | 17490379 | 17490385 | 6 | |
| Deletion | Chr4 | 18036757 | 18036759 | 2 | |
| Deletion | Chr8 | 19173473 | 19173475 | 2 | |
| Deletion | Chr3 | 19455637 | 19455890 | 253 | |
| Deletion | Chr8 | 21001308 | 21001312 | 4 | |
| Deletion | Chr1 | 22353279 | 22353280 | 1 | |
| Deletion | Chr7 | 23113417 | 23113420 | 3 | LOC_Os07g38480 |
| Deletion | Chr7 | 24226206 | 24226231 | 25 | |
| Deletion | Chr5 | 27217752 | 27217755 | 3 | |
| Deletion | Chr3 | 34146411 | 34146412 | 1 | |
| Deletion | Chr1 | 40448182 | 40448189 | 7 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 20126232 | 20126233 | 2 | |
| Insertion | Chr11 | 27282741 | 27282742 | 2 | LOC_Os11g45090 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 5268966 | 5306680 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 6365512 | Chr4 | 16098217 | |
| Translocation | Chr11 | 13147374 | Chr5 | 14831586 | LOC_Os11g22850 |
