Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3311-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3311-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 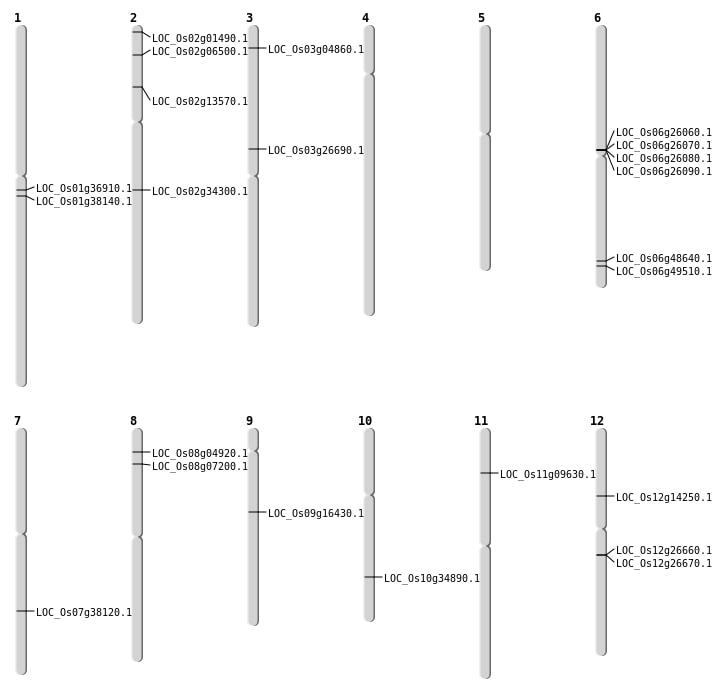
|
| Variant Information |
|---|
Single base substitutions: 67
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15612443 | G-A | HET | ||
| SBS | Chr1 | 1652802 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 18197278 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 37860530 | G-A | HET | INTRON | |
| SBS | Chr1 | 411410 | T-C | HET | ||
| SBS | Chr1 | 43140227 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 11429641 | G-A | Homo | ||
| SBS | Chr10 | 17568036 | G-A | HET | ||
| SBS | Chr10 | 19769132 | C-T | HET | ||
| SBS | Chr10 | 9272945 | A-T | HET | ||
| SBS | Chr10 | 9272946 | C-T | HET | ||
| SBS | Chr11 | 1062294 | A-C | Homo | ||
| SBS | Chr11 | 1062294 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 10713474 | T-G | Homo | ||
| SBS | Chr11 | 20095017 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 23223796 | T-A | Homo | ||
| SBS | Chr11 | 26150586 | A-G | HET | INTRON | |
| SBS | Chr11 | 26150587 | G-A | HET | INTRON | |
| SBS | Chr11 | 5149227 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g09630 |
| SBS | Chr12 | 15907003 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 17127908 | G-A | HET | ||
| SBS | Chr12 | 20223113 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 21775916 | C-A | Homo | ||
| SBS | Chr12 | 4576400 | G-A | HET | ||
| SBS | Chr12 | 8103762 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g14250 |
| SBS | Chr12 | 8169001 | C-T | HET | ||
| SBS | Chr2 | 1492075 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 15082260 | G-T | Homo | ||
| SBS | Chr2 | 20526845 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g34300 |
| SBS | Chr2 | 24910501 | G-A | HET | ||
| SBS | Chr2 | 284955 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g01490 |
| SBS | Chr2 | 32525034 | A-G | HET | ||
| SBS | Chr2 | 35739745 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 8276402 | A-G | HET | ||
| SBS | Chr2 | 9000566 | T-C | HET | ||
| SBS | Chr2 | 9562838 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 22080501 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 23418959 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 24329132 | C-T | Homo | ||
| SBS | Chr3 | 24329132 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 35282164 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 925479 | T-A | HET | ||
| SBS | Chr4 | 21531164 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 2361885 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 24150069 | C-T | Homo | ||
| SBS | Chr4 | 3113116 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 4495336 | C-T | Homo | ||
| SBS | Chr4 | 4495336 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 5189162 | C-T | Homo | ||
| SBS | Chr4 | 5189162 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 17301474 | A-G | HET | ||
| SBS | Chr5 | 4845815 | G-A | HET | ||
| SBS | Chr6 | 19860051 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 23934985 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 29862591 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 1430817 | C-A | HET | ||
| SBS | Chr7 | 19788454 | G-A | HET | ||
| SBS | Chr7 | 21145897 | G-C | HET | UTR_5_PRIME | |
| SBS | Chr8 | 10887889 | T-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 21167471 | G-A | HET | ||
| SBS | Chr8 | 27831968 | G-A | HET | ||
| SBS | Chr8 | 4773395 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 4773396 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 6687178 | A-G | HET | ||
| SBS | Chr9 | 1412031 | G-A | HET | ||
| SBS | Chr9 | 16551172 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 4091276 | C-T | HET | INTRON |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 657341 | 657343 | 2 | |
| Deletion | Chr3 | 2322188 | 2322189 | 1 | LOC_Os03g04860 |
| Deletion | Chr7 | 8519584 | 8519586 | 2 | |
| Deletion | Chr7 | 9446881 | 9446886 | 5 | |
| Deletion | Chr10 | 11547349 | 11547353 | 4 | |
| Deletion | Chr5 | 13648799 | 13648801 | 2 | |
| Deletion | Chr7 | 14152900 | 14152901 | 1 | |
| Deletion | Chr6 | 15247001 | 15271000 | 23999 | 4 |
| Deletion | Chr2 | 15266974 | 15266977 | 3 | |
| Deletion | Chr12 | 15622001 | 15635000 | 12999 | 2 |
| Deletion | Chr8 | 15843495 | 15843500 | 5 | |
| Deletion | Chr12 | 15976899 | 15976905 | 6 | |
| Deletion | Chr10 | 17584101 | 17584119 | 18 | |
| Deletion | Chr8 | 18552494 | 18552495 | 1 | 2 |
| Deletion | Chr10 | 18627946 | 18627952 | 6 | LOC_Os10g34890 |
| Deletion | Chr5 | 18895369 | 18895374 | 5 | |
| Deletion | Chr7 | 19826377 | 19826379 | 2 | |
| Deletion | Chr2 | 20302149 | 20302150 | 1 | |
| Deletion | Chr1 | 20568250 | 20568270 | 20 | LOC_Os01g36910 |
| Deletion | Chr1 | 21356613 | 21356615 | 2 | LOC_Os01g38140 |
| Deletion | Chr6 | 21890412 | 21890417 | 5 | |
| Deletion | Chr3 | 26603520 | 26603521 | 1 | |
| Deletion | Chr1 | 29702117 | 29702122 | 5 | |
| Deletion | Chr1 | 34117933 | 34117935 | 2 | |
| Deletion | Chr1 | 41782582 | 41782589 | 7 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 12030010 | 12030010 | 1 | |
| Insertion | Chr2 | 9682858 | 9682858 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 10043623 | 10070079 | LOC_Os09g16430 |
| Inversion | Chr1 | 22238323 | 22242058 | |
| Inversion | Chr1 | 22238335 | 22242063 | |
| Inversion | Chr6 | 29424132 | 29993996 | 2 |
| Inversion | Chr6 | 29424139 | 29994001 | 2 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 1003520 | Chr3 | 15238126 | LOC_Os03g26690 |
| Translocation | Chr2 | 3249550 | Chr1 | 35549254 | LOC_Os02g06500 |
| Translocation | Chr2 | 3249552 | Chr1 | 35550127 | LOC_Os02g06500 |
| Translocation | Chr8 | 4019874 | Chr7 | 22860005 | 2 |
| Translocation | Chr8 | 4019875 | Chr7 | 22859719 | 2 |
| Translocation | Chr10 | 8059531 | Chr2 | 7279936 | 2 |
| Translocation | Chr10 | 8082064 | Chr2 | 7279933 | 2 |
| Translocation | Chr9 | 20666147 | Chr8 | 2513188 | LOC_Os08g04920 |
