Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3364-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3364-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 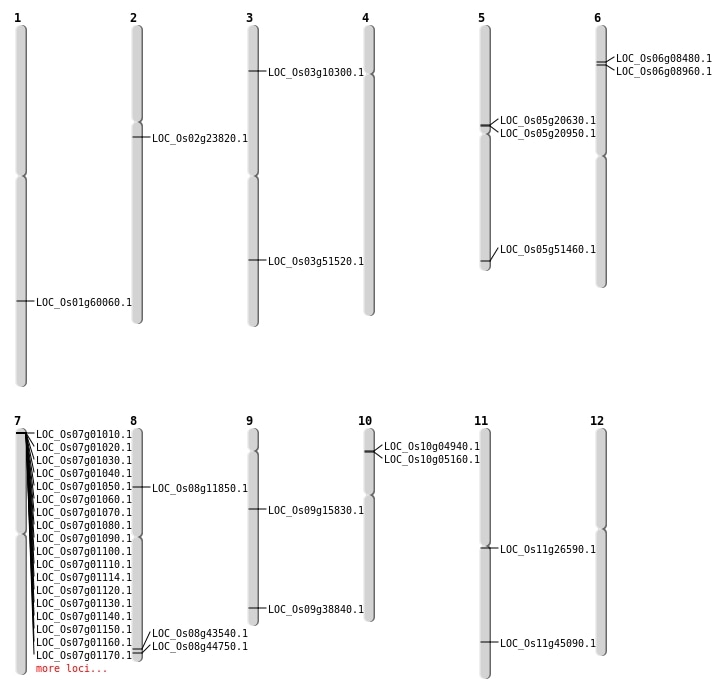
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19608844 | A-G | Homo | ||
| SBS | Chr1 | 34740550 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g60060 |
| SBS | Chr1 | 35731286 | C-T | Homo | ||
| SBS | Chr1 | 36413971 | T-A | HET | ||
| SBS | Chr1 | 36413972 | C-T | HET | ||
| SBS | Chr1 | 40678994 | T-C | HET | ||
| SBS | Chr1 | 814129 | C-T | HET | ||
| SBS | Chr10 | 16534 | C-T | HET | ||
| SBS | Chr10 | 6365661 | G-T | HET | ||
| SBS | Chr10 | 9766594 | C-T | HET | ||
| SBS | Chr11 | 15229491 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g26590 |
| SBS | Chr11 | 21431632 | C-T | HET | ||
| SBS | Chr12 | 25455213 | G-A | Homo | ||
| SBS | Chr12 | 27180281 | C-T | HET | ||
| SBS | Chr12 | 7301899 | G-A | Homo | ||
| SBS | Chr2 | 30497438 | C-T | HET | ||
| SBS | Chr2 | 6615250 | T-A | HET | ||
| SBS | Chr2 | 8096534 | A-T | HET | ||
| SBS | Chr3 | 29468234 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g51520 |
| SBS | Chr3 | 5254432 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g10300 |
| SBS | Chr3 | 7179528 | C-T | HET | ||
| SBS | Chr4 | 6809474 | G-A | HET | ||
| SBS | Chr5 | 12320098 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g20950 |
| SBS | Chr5 | 20874200 | T-C | HET | ||
| SBS | Chr5 | 20878085 | G-A | HET | ||
| SBS | Chr5 | 21132329 | G-A | HET | ||
| SBS | Chr5 | 28881434 | G-A | Homo | ||
| SBS | Chr5 | 29503581 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g51460 |
| SBS | Chr5 | 8345233 | T-C | HET | ||
| SBS | Chr6 | 11164027 | C-T | Homo | ||
| SBS | Chr6 | 13451574 | C-T | HET | ||
| SBS | Chr6 | 21359061 | G-A | HET | ||
| SBS | Chr6 | 26621253 | G-A | HET | ||
| SBS | Chr6 | 29486981 | T-A | HET | ||
| SBS | Chr6 | 326151 | C-T | HET | ||
| SBS | Chr6 | 4483918 | T-C | HET | ||
| SBS | Chr6 | 8284243 | G-A | HET | ||
| SBS | Chr7 | 19215266 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g32310 |
| SBS | Chr7 | 21565052 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36060 |
| SBS | Chr7 | 22457573 | C-T | HET | ||
| SBS | Chr7 | 24005781 | C-A | HET | ||
| SBS | Chr7 | 3987355 | T-A | Homo | ||
| SBS | Chr7 | 578085 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g01940 |
| SBS | Chr8 | 14713025 | A-G | HET | ||
| SBS | Chr8 | 6950254 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g11850 |
| SBS | Chr9 | 19252001 | C-T | HET |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 1 | 153000 | 152999 | 25 |
| Deletion | Chr7 | 397001 | 488000 | 90999 | 17 |
| Deletion | Chr1 | 5483190 | 5483191 | 1 | |
| Deletion | Chr4 | 6222618 | 6222625 | 7 | |
| Deletion | Chr3 | 8190469 | 8190475 | 6 | |
| Deletion | Chr11 | 9739486 | 9739489 | 3 | |
| Deletion | Chr8 | 10480847 | 10480849 | 2 | |
| Deletion | Chr6 | 11163272 | 11163283 | 11 | |
| Deletion | Chr3 | 14267157 | 14267160 | 3 | |
| Deletion | Chr4 | 14901222 | 14901235 | 13 | |
| Deletion | Chr2 | 16030669 | 16030671 | 2 | |
| Deletion | Chr3 | 16410504 | 16410513 | 9 | |
| Deletion | Chr3 | 17390556 | 17390563 | 7 | |
| Deletion | Chr1 | 17972626 | 17972633 | 7 | |
| Deletion | Chr11 | 19498192 | 19498193 | 1 | |
| Deletion | Chr9 | 22312011 | 22312021 | 10 | LOC_Os09g38840 |
| Deletion | Chr9 | 22694988 | 22694997 | 9 | |
| Deletion | Chr3 | 23685734 | 23685737 | 3 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 19416072 | 19416072 | 1 | |
| Insertion | Chr11 | 19416435 | 19416436 | 2 | |
| Insertion | Chr11 | 8300268 | 8300269 | 2 | |
| Insertion | Chr12 | 8264472 | 8264474 | 3 | |
| Insertion | Chr2 | 13709743 | 13709743 | 1 | LOC_Os02g23820 |
| Insertion | Chr4 | 23301401 | 23301402 | 2 | |
| Insertion | Chr4 | 6331862 | 6331862 | 1 | |
| Insertion | Chr6 | 8656151 | 8656174 | 24 |
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 2412331 | 2519833 | 2 |
| Inversion | Chr6 | 4181437 | 4523017 | 2 |
| Inversion | Chr6 | 4181447 | 4524642 | 2 |
| Inversion | Chr6 | 4181448 | 4523020 | 2 |
| Inversion | Chr9 | 9661948 | 9954134 | LOC_Os09g15830 |
| Inversion | Chr8 | 27300291 | 27535885 | 2 |
| Inversion | Chr8 | 27300321 | 27535893 | 2 |
| Inversion | Chr8 | 28120780 | 28203172 | LOC_Os08g44750 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 4244177 | Chr2 | 6526413 | |
| Translocation | Chr11 | 15950035 | Chr1 | 14847997 | |
| Translocation | Chr12 | 18707492 | Chr5 | 12102772 | 2 |
| Translocation | Chr5 | 19391900 | Chr2 | 17106255 | |
| Translocation | Chr8 | 19479857 | Chr4 | 35498773 | |
| Translocation | Chr11 | 22995419 | Chr7 | 21960411 | |
| Translocation | Chr11 | 27282751 | Chr10 | 17466608 | LOC_Os11g45090 |
