Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3373-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3373-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 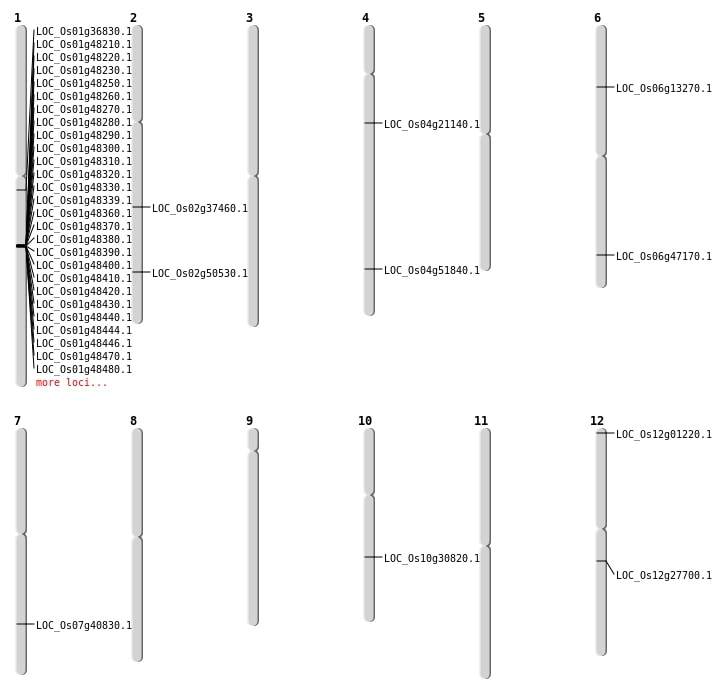
|
| Variant Information |
|---|
Single base substitutions: 52
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11588451 | T-G | HET | ||
| SBS | Chr1 | 12686863 | A-G | HET | ||
| SBS | Chr1 | 15873160 | G-A | Homo | ||
| SBS | Chr1 | 20487898 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g36830 |
| SBS | Chr1 | 24321485 | T-C | HET | ||
| SBS | Chr1 | 24759701 | G-C | HET | ||
| SBS | Chr1 | 24759702 | G-A | HET | ||
| SBS | Chr1 | 28174436 | T-C | HET | ||
| SBS | Chr1 | 41239313 | T-A | HET | ||
| SBS | Chr10 | 15239309 | T-A | HET | ||
| SBS | Chr10 | 15516961 | T-G | HET | ||
| SBS | Chr10 | 383740 | T-C | HET | ||
| SBS | Chr10 | 803716 | A-G | HET | ||
| SBS | Chr11 | 20395570 | C-T | HET | ||
| SBS | Chr11 | 24928136 | C-T | HET | ||
| SBS | Chr11 | 24928137 | A-T | HET | ||
| SBS | Chr12 | 16336103 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g27700 |
| SBS | Chr12 | 17277408 | C-T | HET | ||
| SBS | Chr12 | 17595713 | G-C | HET | ||
| SBS | Chr2 | 21942566 | C-T | HET | ||
| SBS | Chr2 | 22615005 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g37460 |
| SBS | Chr2 | 30856384 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g50530 |
| SBS | Chr2 | 31293070 | C-T | HET | ||
| SBS | Chr2 | 3525358 | T-G | HET | ||
| SBS | Chr3 | 15535156 | C-G | HET | ||
| SBS | Chr3 | 16212750 | T-A | HET | ||
| SBS | Chr3 | 546001 | C-T | HET | ||
| SBS | Chr4 | 11901980 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g21140 |
| SBS | Chr4 | 23427262 | T-A | Homo | ||
| SBS | Chr4 | 30557909 | A-G | HET | ||
| SBS | Chr4 | 30745510 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g51840 |
| SBS | Chr4 | 34886243 | G-T | HET | ||
| SBS | Chr5 | 11730772 | C-T | HET | ||
| SBS | Chr5 | 14641284 | T-G | HET | ||
| SBS | Chr5 | 15697565 | C-T | HET | ||
| SBS | Chr5 | 20542035 | T-C | HET | ||
| SBS | Chr5 | 417247 | G-T | HET | ||
| SBS | Chr5 | 417248 | A-T | HET | ||
| SBS | Chr6 | 15365552 | G-C | HET | ||
| SBS | Chr6 | 20120902 | T-G | HET | ||
| SBS | Chr6 | 28603165 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g47170 |
| SBS | Chr6 | 28603166 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g47170 |
| SBS | Chr7 | 27991206 | C-T | HET | ||
| SBS | Chr7 | 4347026 | T-A | HET | ||
| SBS | Chr7 | 6022004 | G-A | HET | ||
| SBS | Chr8 | 11338531 | A-G | HET | ||
| SBS | Chr8 | 11524012 | C-T | HET | ||
| SBS | Chr8 | 17555664 | C-T | HET | ||
| SBS | Chr8 | 21312823 | C-T | HET | ||
| SBS | Chr8 | 23815433 | C-T | Homo | ||
| SBS | Chr8 | 6188870 | G-T | Homo | ||
| SBS | Chr9 | 22007946 | T-C | Homo |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 47399 | 47407 | 8 | |
| Deletion | Chr8 | 508858 | 508869 | 11 | |
| Deletion | Chr6 | 7301224 | 7301225 | 1 | LOC_Os06g13270 |
| Deletion | Chr9 | 8164245 | 8164246 | 1 | |
| Deletion | Chr9 | 10066747 | 10066749 | 2 | |
| Deletion | Chr2 | 10140847 | 10140849 | 2 | |
| Deletion | Chr11 | 10978987 | 10978994 | 7 | |
| Deletion | Chr12 | 15989000 | 15989008 | 8 | |
| Deletion | Chr10 | 16052745 | 16052784 | 39 | LOC_Os10g30820 |
| Deletion | Chr7 | 18376400 | 18376401 | 1 | |
| Deletion | Chr4 | 20756980 | 20756981 | 1 | |
| Deletion | Chr12 | 26528298 | 26528308 | 10 | |
| Deletion | Chr1 | 27613001 | 27807000 | 193999 | 28 |
No Insertion
No Inversion
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 130894 | Chr4 | 23247558 | LOC_Os12g01220 |
| Translocation | Chr12 | 130903 | Chr4 | 23268459 | LOC_Os12g01220 |
| Translocation | Chr10 | 4000966 | Chr7 | 3512212 | |
| Translocation | Chr8 | 17449731 | Chr5 | 27670551 | |
| Translocation | Chr7 | 24466975 | Chr6 | 28709135 | LOC_Os07g40830 |
| Translocation | Chr7 | 25127591 | Chr2 | 21836310 |
