Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3375-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3375-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 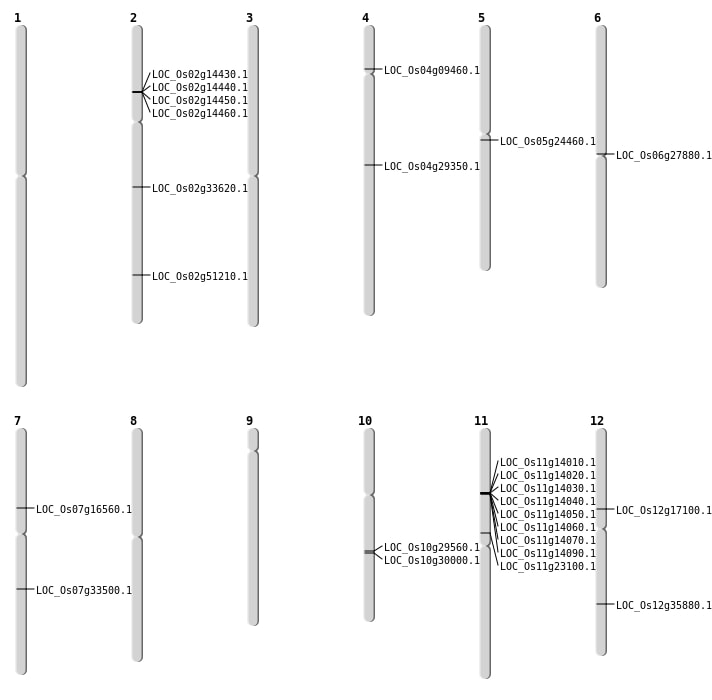
|
| Variant Information |
|---|
Single base substitutions: 49
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15964517 | G-A | HET | ||
| SBS | Chr1 | 3357823 | C-T | HET | ||
| SBS | Chr1 | 37925468 | G-T | HET | ||
| SBS | Chr1 | 39479532 | C-T | HET | ||
| SBS | Chr10 | 15367154 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g29560 |
| SBS | Chr10 | 15572470 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g30000 |
| SBS | Chr10 | 4777203 | A-G | HET | ||
| SBS | Chr11 | 13294811 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g23100 |
| SBS | Chr11 | 17903727 | G-C | HET | ||
| SBS | Chr11 | 26129860 | G-A | HET | ||
| SBS | Chr11 | 7953533 | T-A | HET | ||
| SBS | Chr11 | 9043034 | A-G | HET | ||
| SBS | Chr11 | 9854716 | G-T | HET | ||
| SBS | Chr12 | 21864660 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g35880 |
| SBS | Chr12 | 22335654 | G-C | HET | ||
| SBS | Chr12 | 9793157 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g17100 |
| SBS | Chr2 | 20018994 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g33620 |
| SBS | Chr2 | 35585473 | G-A | Homo | ||
| SBS | Chr2 | 9210795 | C-G | HET | ||
| SBS | Chr3 | 261056 | C-T | Homo | ||
| SBS | Chr3 | 36410328 | A-G | HET | ||
| SBS | Chr3 | 9556922 | A-C | Homo | ||
| SBS | Chr4 | 10288344 | G-A | HET | ||
| SBS | Chr4 | 29153756 | G-A | HET | ||
| SBS | Chr4 | 5066603 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g09460 |
| SBS | Chr4 | 8829256 | C-A | HET | ||
| SBS | Chr5 | 14120674 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g24460 |
| SBS | Chr5 | 14878393 | C-G | HET | ||
| SBS | Chr5 | 15898557 | C-G | HET | ||
| SBS | Chr5 | 27877184 | T-C | HET | ||
| SBS | Chr5 | 29783043 | G-A | HET | ||
| SBS | Chr6 | 12672722 | C-T | Homo | ||
| SBS | Chr6 | 14712545 | A-T | Homo | ||
| SBS | Chr6 | 15801515 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g27880 |
| SBS | Chr6 | 15843345 | G-T | Homo | ||
| SBS | Chr6 | 17652576 | T-A | Homo | ||
| SBS | Chr6 | 27956423 | T-C | Homo | ||
| SBS | Chr6 | 7982698 | C-T | Homo | ||
| SBS | Chr7 | 17701032 | G-A | HET | ||
| SBS | Chr7 | 20023794 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g33500 |
| SBS | Chr7 | 20401329 | C-T | Homo | ||
| SBS | Chr7 | 22079592 | C-T | Homo | ||
| SBS | Chr7 | 24100124 | T-A | HET | ||
| SBS | Chr7 | 8437178 | G-T | Homo | ||
| SBS | Chr8 | 6338575 | T-C | HET | ||
| SBS | Chr8 | 7539273 | C-T | HET | ||
| SBS | Chr9 | 16376155 | G-A | HET | ||
| SBS | Chr9 | 7847293 | C-T | Homo | ||
| SBS | Chr9 | 9998858 | G-A | HET |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 603687 | 603688 | 1 | |
| Deletion | Chr6 | 5381965 | 5381966 | 1 | |
| Deletion | Chr11 | 7775001 | 7819000 | 43999 | 7 |
| Deletion | Chr2 | 7925001 | 7963000 | 37999 | 4 |
| Deletion | Chr8 | 9140001 | 9148000 | 7999 | |
| Deletion | Chr1 | 10443199 | 10443207 | 8 | |
| Deletion | Chr10 | 12312212 | 12312217 | 5 | |
| Deletion | Chr2 | 19075817 | 19075821 | 4 | |
| Deletion | Chr6 | 20262398 | 20262399 | 1 | |
| Deletion | Chr9 | 21170308 | 21170309 | 1 | |
| Deletion | Chr6 | 25958565 | 25958570 | 5 | |
| Deletion | Chr1 | 26317392 | 26317410 | 18 | |
| Deletion | Chr2 | 31338308 | 31338311 | 3 | LOC_Os02g51210 |
Insertions: 6
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 9582428 | 9710571 | 2 |
| Inversion | Chr7 | 9582445 | 9710583 | 2 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 6239638 | Chr1 | 11667832 | |
| Translocation | Chr11 | 7842111 | Chr4 | 17430870 | 2 |
| Translocation | Chr12 | 15658464 | Chr8 | 15380492 | |
| Translocation | Chr6 | 17270453 | Chr2 | 22538977 | |
| Translocation | Chr11 | 28093710 | Chr7 | 21747120 |
