Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3387-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3387-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 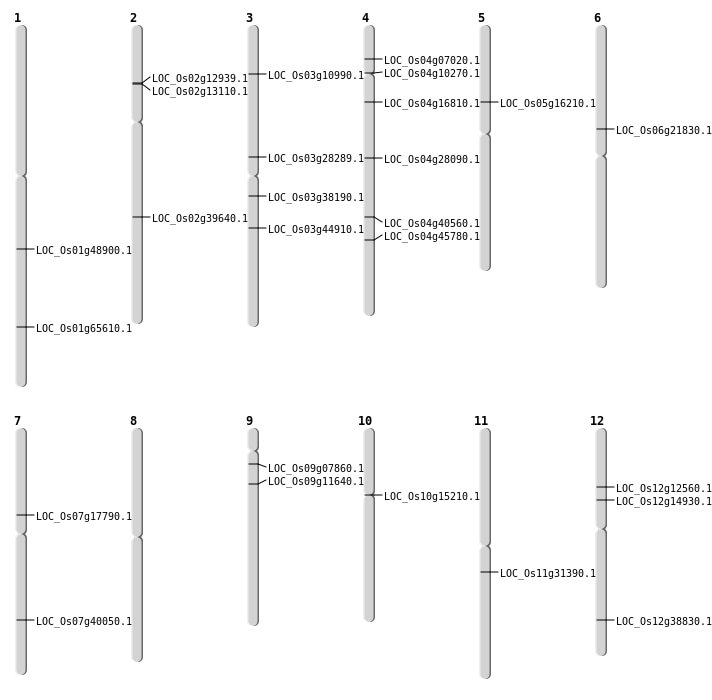
|
| Variant Information |
|---|
Single base substitutions: 59
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14711726 | C-T | HET | ||
| SBS | Chr1 | 15426404 | G-C | HET | ||
| SBS | Chr1 | 20226927 | C-G | HET | ||
| SBS | Chr1 | 22665269 | C-T | HET | ||
| SBS | Chr1 | 27206905 | T-A | HET | ||
| SBS | Chr1 | 35462489 | G-A | HET | ||
| SBS | Chr1 | 40038661 | T-C | HET | ||
| SBS | Chr1 | 8449250 | C-T | HET | ||
| SBS | Chr10 | 13596520 | G-A | HET | ||
| SBS | Chr10 | 3842973 | C-G | HET | ||
| SBS | Chr11 | 14719169 | C-A | HET | ||
| SBS | Chr11 | 16084954 | G-T | HET | ||
| SBS | Chr11 | 18318609 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g31390 |
| SBS | Chr11 | 23899845 | T-A | Homo | ||
| SBS | Chr12 | 11363379 | A-T | Homo | ||
| SBS | Chr12 | 12829338 | G-A | Homo | ||
| SBS | Chr12 | 1547159 | T-C | HET | ||
| SBS | Chr12 | 15723995 | T-C | HET | ||
| SBS | Chr12 | 23860872 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g38830 |
| SBS | Chr12 | 23923602 | T-A | HET | ||
| SBS | Chr2 | 5611364 | A-G | HET | ||
| SBS | Chr3 | 10142804 | A-T | HET | ||
| SBS | Chr3 | 10680431 | A-G | HET | ||
| SBS | Chr3 | 16280520 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g28289 |
| SBS | Chr3 | 19383665 | A-C | HET | ||
| SBS | Chr3 | 21476012 | G-A | Homo | ||
| SBS | Chr3 | 23622225 | C-A | HET | ||
| SBS | Chr3 | 27451574 | C-T | HET | ||
| SBS | Chr3 | 30157618 | C-T | Homo | ||
| SBS | Chr3 | 5648203 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g10990 |
| SBS | Chr4 | 24087813 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g40560 |
| SBS | Chr4 | 27093125 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g45780 |
| SBS | Chr4 | 27160620 | A-G | Homo | ||
| SBS | Chr4 | 34011758 | G-T | HET | ||
| SBS | Chr4 | 7019832 | G-A | HET | ||
| SBS | Chr4 | 9212804 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g16810 |
| SBS | Chr5 | 12804950 | G-A | Homo | ||
| SBS | Chr5 | 19428225 | C-G | HET | ||
| SBS | Chr5 | 23363757 | C-T | HET | ||
| SBS | Chr5 | 5008305 | C-T | HET | ||
| SBS | Chr6 | 10454091 | C-T | HET | ||
| SBS | Chr6 | 12608987 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g21830 |
| SBS | Chr6 | 6332625 | C-A | HET | ||
| SBS | Chr7 | 10145095 | G-T | HET | ||
| SBS | Chr7 | 10524864 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g17790 |
| SBS | Chr7 | 12403177 | C-T | HET | ||
| SBS | Chr7 | 15480822 | G-T | HET | ||
| SBS | Chr7 | 21393133 | C-T | HET | ||
| SBS | Chr7 | 21393135 | A-C | HET | ||
| SBS | Chr7 | 22988869 | T-A | HET | ||
| SBS | Chr7 | 26844200 | G-A | Homo | ||
| SBS | Chr7 | 3519047 | C-A | HET | ||
| SBS | Chr7 | 8563563 | A-G | HET | ||
| SBS | Chr7 | 898606 | T-C | HET | ||
| SBS | Chr8 | 12628745 | C-G | HET | ||
| SBS | Chr8 | 15545097 | C-T | HET | ||
| SBS | Chr8 | 3595828 | A-G | Homo | ||
| SBS | Chr9 | 3997712 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07860 |
| SBS | Chr9 | 6498645 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g11640 |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 719532 | 719533 | 1 | |
| Deletion | Chr11 | 2971334 | 2971338 | 4 | |
| Deletion | Chr4 | 3735891 | 3735892 | 1 | LOC_Os04g07020 |
| Deletion | Chr4 | 5548338 | 5548339 | 1 | LOC_Os04g10270 |
| Deletion | Chr12 | 6912403 | 6912407 | 4 | LOC_Os12g12560 |
| Deletion | Chr9 | 7253929 | 7253998 | 69 | |
| Deletion | Chr11 | 12790295 | 12790296 | 1 | |
| Deletion | Chr10 | 13815480 | 13815485 | 5 | |
| Deletion | Chr11 | 19154339 | 19154349 | 10 | |
| Deletion | Chr9 | 20993213 | 20993214 | 1 | |
| Deletion | Chr8 | 23172154 | 23172156 | 2 | |
| Deletion | Chr3 | 23634021 | 23634023 | 2 | |
| Deletion | Chr1 | 23718322 | 23718328 | 6 | |
| Deletion | Chr2 | 23915001 | 23940000 | 24999 | LOC_Os02g39640 |
| Deletion | Chr7 | 24043380 | 24043392 | 12 | LOC_Os07g40050 |
| Deletion | Chr7 | 28186419 | 28186420 | 1 | |
| Deletion | Chr3 | 32554164 | 32554166 | 2 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 8177215 | 8177215 | 1 | |
| Insertion | Chr8 | 11819534 | 11819534 | 1 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 6831603 | 6969729 | 2 |
| Inversion | Chr2 | 6831607 | 6969741 | 2 |
| Inversion | Chr3 | 20672733 | 21208542 | 2 |
| Inversion | Chr3 | 20672735 | 21208549 | 2 |
| Inversion | Chr2 | 23232144 | 23232312 | |
| Inversion | Chr1 | 28058486 | 28058769 | LOC_Os01g48900 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 1609517 | Chr8 | 2830837 | |
| Translocation | Chr10 | 1609838 | Chr8 | 2830861 | |
| Translocation | Chr11 | 5025428 | Chr8 | 3815240 | |
| Translocation | Chr3 | 5504283 | Chr1 | 38091882 | 2 |
| Translocation | Chr10 | 8050300 | Chr8 | 18696417 | LOC_Os10g15210 |
| Translocation | Chr12 | 8540937 | Chr1 | 39127625 | LOC_Os12g14930 |
| Translocation | Chr5 | 9171432 | Chr3 | 25355901 | 2 |
| Translocation | Chr4 | 16584622 | Chr2 | 20382715 | LOC_Os04g28090 |
| Translocation | Chr6 | 20576932 | Chr4 | 9404447 |
