Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN347-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN347-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 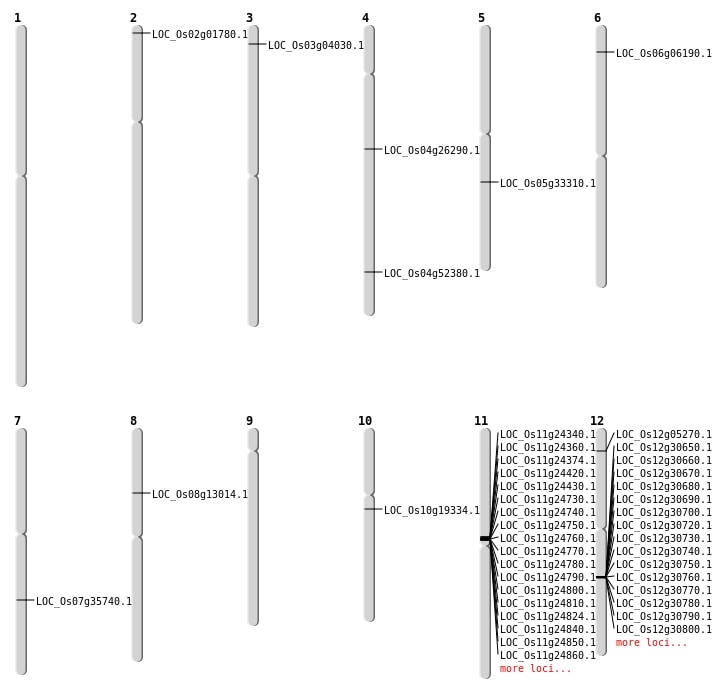
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14148653 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 5735058 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1071114 | A-T | HET | INTRON | |
| SBS | Chr10 | 3379884 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 9766095 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 12901836 | G-T | HET | INTRON | |
| SBS | Chr11 | 23961338 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 11253480 | A-G | HOMO | INTRON | |
| SBS | Chr12 | 23437937 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 2346729 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g05270 |
| SBS | Chr2 | 10199560 | T-A | HET | INTRON | |
| SBS | Chr2 | 11385050 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 21796919 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 21796920 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 35622642 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 1401394 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 14094468 | G-T | HET | UTR_5_PRIME | |
| SBS | Chr3 | 1838424 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g04030 |
| SBS | Chr3 | 20111963 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 27426126 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 27524491 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 11644832 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 15325223 | C-A | HET | STOP_GAINED | LOC_Os04g26290 |
| SBS | Chr4 | 19045630 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 4979170 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 18217872 | T-C | HET | INTRON | |
| SBS | Chr5 | 19527194 | A-T | HET | STOP_GAINED | LOC_Os05g33310 |
| SBS | Chr5 | 20369899 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 23033123 | G-A | HET | INTRON | |
| SBS | Chr5 | 24854815 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 27873475 | C-A | HET | INTRON | |
| SBS | Chr6 | 10852504 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 20211010 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 29360819 | T-A | HET | INTRON | |
| SBS | Chr7 | 18745419 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 21408732 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g35740 |
| SBS | Chr7 | 9682502 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 27690350 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 15375949 | C-T | HOMO | INTERGENIC |
Deletions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 430513 | 430528 | 15 | LOC_Os02g01780 |
| Deletion | Chr3 | 2653037 | 2653038 | 1 | |
| Deletion | Chr6 | 2867578 | 2867589 | 11 | LOC_Os06g06190 |
| Deletion | Chr1 | 6568349 | 6568351 | 2 | |
| Deletion | Chr11 | 11623989 | 11623993 | 4 | |
| Deletion | Chr11 | 13863001 | 13932000 | 69000 | 5 |
| Deletion | Chr11 | 14080338 | 14788334 | 707996 | 107 |
| Deletion | Chr9 | 15342427 | 15342428 | 1 | |
| Deletion | Chr12 | 18414001 | 18578000 | 164000 | 23 |
| Deletion | Chr2 | 20131160 | 20131161 | 1 | |
| Deletion | Chr4 | 29212866 | 29212867 | 1 | |
| Deletion | Chr6 | 29374568 | 29374580 | 12 | |
| Deletion | Chr3 | 32577821 | 32577850 | 29 | |
| Deletion | Chr1 | 42722841 | 42722842 | 1 |
Insertions: 6
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 12838241 | 14115710 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 7731058 | Chr5 | 4615621 | LOC_Os08g13014 |
| Translocation | Chr10 | 9867991 | Chr4 | 31119077 | LOC_Os10g19334 |
| Translocation | Chr10 | 9867993 | Chr4 | 31121082 | 2 |
