Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3479-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3479-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 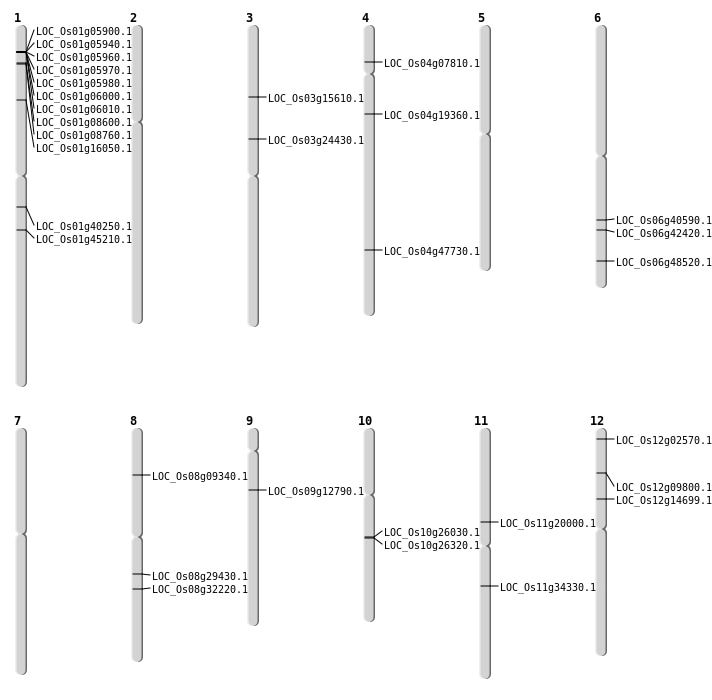
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17520922 | C-A | HET | ||
| SBS | Chr1 | 25654374 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g45210 |
| SBS | Chr1 | 40849178 | C-G | HET | ||
| SBS | Chr1 | 9785559 | G-A | HET | ||
| SBS | Chr10 | 13474857 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g26030 |
| SBS | Chr10 | 13486323 | G-T | HET | ||
| SBS | Chr10 | 14086453 | G-T | HET | ||
| SBS | Chr10 | 2064914 | T-A | HET | ||
| SBS | Chr10 | 4999740 | G-A | HET | ||
| SBS | Chr11 | 11460374 | C-T | Homo | ||
| SBS | Chr11 | 17722807 | A-C | Homo | ||
| SBS | Chr11 | 26718397 | G-A | HET | ||
| SBS | Chr12 | 23089165 | A-G | HET | ||
| SBS | Chr12 | 5194413 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g09800 |
| SBS | Chr12 | 8131743 | A-C | HET | ||
| SBS | Chr12 | 8411404 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g14699 |
| SBS | Chr3 | 13916470 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g24430 |
| SBS | Chr3 | 26752743 | G-T | Homo | ||
| SBS | Chr3 | 8614500 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g15610 |
| SBS | Chr4 | 10775238 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g19360 |
| SBS | Chr4 | 24801888 | A-G | Homo | ||
| SBS | Chr4 | 27094241 | C-A | Homo | ||
| SBS | Chr4 | 28320812 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g47730 |
| SBS | Chr4 | 4144665 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g07810 |
| SBS | Chr6 | 12681131 | G-A | HET | ||
| SBS | Chr6 | 1623854 | T-C | HET | ||
| SBS | Chr6 | 24602452 | A-G | HET | ||
| SBS | Chr6 | 27744153 | A-T | Homo | ||
| SBS | Chr6 | 29025126 | C-A | Homo | ||
| SBS | Chr6 | 29354866 | A-T | Homo | STOP_GAINED | LOC_Os06g48520 |
| SBS | Chr6 | 29354867 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g48520 |
| SBS | Chr6 | 29354868 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g48520 |
| SBS | Chr7 | 10440402 | T-C | Homo | ||
| SBS | Chr8 | 18045086 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g29430 |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 2830001 | 2874000 | 43999 | 7 |
| Deletion | Chr7 | 7063953 | 7063970 | 17 | |
| Deletion | Chr9 | 7339645 | 7339655 | 10 | LOC_Os09g12790 |
| Deletion | Chr6 | 7607778 | 7607779 | 1 | |
| Deletion | Chr2 | 8430722 | 8430732 | 10 | |
| Deletion | Chr12 | 10497690 | 10497691 | 1 | |
| Deletion | Chr7 | 10543985 | 10544012 | 27 | |
| Deletion | Chr8 | 10746971 | 10746980 | 9 | |
| Deletion | Chr10 | 11067528 | 11067534 | 6 | |
| Deletion | Chr7 | 11136195 | 11136206 | 11 | |
| Deletion | Chr10 | 13646720 | 13646722 | 2 | LOC_Os10g26320 |
| Deletion | Chr8 | 17192637 | 17192638 | 1 | |
| Deletion | Chr1 | 22714001 | 22728000 | 13999 | LOC_Os01g40250 |
| Deletion | Chr6 | 24537399 | 24537400 | 1 | |
| Deletion | Chr6 | 25495452 | 25495475 | 23 | LOC_Os06g42420 |
| Deletion | Chr1 | 25933066 | 25933080 | 14 | |
| Deletion | Chr2 | 29706893 | 29706895 | 2 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 9033670 | 9033670 | 1 | LOC_Os01g16050 |
| Insertion | Chr10 | 1596388 | 1596390 | 3 | |
| Insertion | Chr3 | 1909278 | 1909279 | 2 | |
| Insertion | Chr6 | 11823715 | 11823715 | 1 | |
| Insertion | Chr9 | 4081073 | 4081073 | 1 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 4283260 | 4395954 | 2 |
| Inversion | Chr8 | 4738129 | 5419138 | 2 |
| Inversion | Chr8 | 4738139 | 5419138 | 2 |
| Inversion | Chr6 | 13329822 | 13672488 | LOC_Os06g22850 |
| Inversion | Chr6 | 13329826 | 13672500 | LOC_Os06g22850 |
| Inversion | Chr6 | 24199002 | 24213784 | LOC_Os06g40590 |
| Inversion | Chr6 | 24199005 | 24213784 | LOC_Os06g40590 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 888362 | Chr7 | 21020817 | LOC_Os12g02570 |
| Translocation | Chr4 | 6335793 | Chr1 | 24632607 | |
| Translocation | Chr11 | 10569843 | Chr9 | 7975560 | |
| Translocation | Chr11 | 11508165 | Chr8 | 19974189 | 2 |
| Translocation | Chr11 | 20126119 | Chr3 | 8260847 | LOC_Os11g34330 |
