Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3514-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3514-S Alignment File |
| Seed Availability | No |
| Mapping (Hover to Zoom-In) | 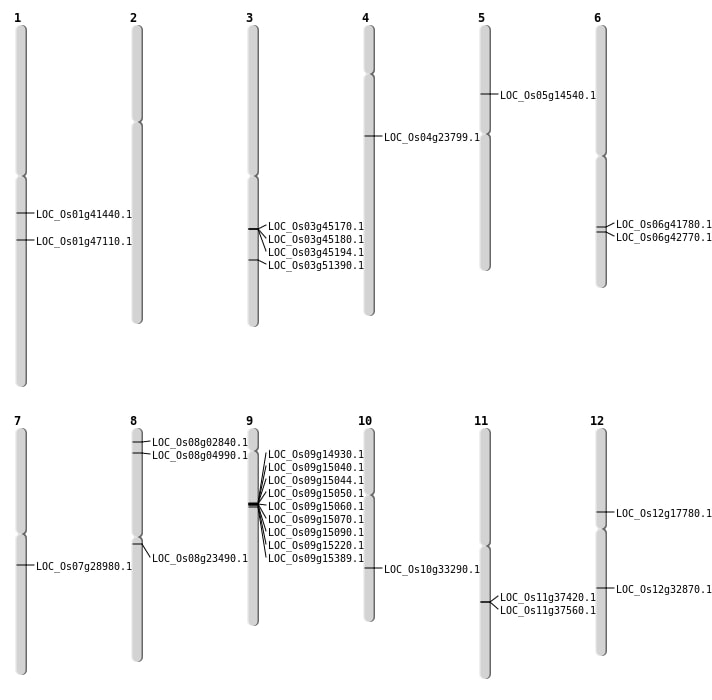
|
| Variant Information |
|---|
Single base substitutions: 57
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22892462 | G-A | Homo | ||
| SBS | Chr1 | 24521704 | T-C | Homo | ||
| SBS | Chr1 | 26916246 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g47110 |
| SBS | Chr1 | 26931193 | C-T | Homo | ||
| SBS | Chr1 | 36452460 | T-C | HET | ||
| SBS | Chr1 | 5547274 | C-A | HET | ||
| SBS | Chr1 | 5547276 | C-T | HET | ||
| SBS | Chr10 | 17487827 | C-A | HET | STOP_GAINED | LOC_Os10g33290 |
| SBS | Chr10 | 18916786 | G-A | HET | ||
| SBS | Chr10 | 20407015 | C-A | HET | ||
| SBS | Chr10 | 9582027 | G-T | HET | ||
| SBS | Chr11 | 22021445 | G-A | HET | ||
| SBS | Chr11 | 9328603 | C-A | HET | ||
| SBS | Chr12 | 10176459 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g17780 |
| SBS | Chr12 | 19574981 | A-T | HET | ||
| SBS | Chr12 | 19865987 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g32870 |
| SBS | Chr12 | 21764826 | T-C | HET | ||
| SBS | Chr12 | 26191153 | C-T | HET | ||
| SBS | Chr12 | 26728214 | C-T | HET | ||
| SBS | Chr12 | 5053394 | A-T | HET | ||
| SBS | Chr12 | 5754440 | T-A | HET | ||
| SBS | Chr2 | 10812792 | C-A | HET | ||
| SBS | Chr2 | 29614320 | C-T | HET | ||
| SBS | Chr2 | 3619515 | G-T | HET | ||
| SBS | Chr2 | 535446 | T-C | HET | ||
| SBS | Chr2 | 7830411 | C-T | HET | ||
| SBS | Chr2 | 8966111 | C-T | HET | ||
| SBS | Chr3 | 14139890 | C-T | HET | ||
| SBS | Chr3 | 17376274 | G-T | Homo | ||
| SBS | Chr3 | 25559322 | C-T | Homo | ||
| SBS | Chr3 | 29141056 | A-T | Homo | ||
| SBS | Chr3 | 5900074 | T-C | HET | ||
| SBS | Chr4 | 10741855 | G-A | Homo | ||
| SBS | Chr4 | 16800770 | G-A | HET | ||
| SBS | Chr4 | 17339276 | C-T | Homo | ||
| SBS | Chr4 | 25261197 | G-T | HET | ||
| SBS | Chr4 | 31114238 | A-G | HET | ||
| SBS | Chr5 | 10229119 | T-C | HET | ||
| SBS | Chr5 | 10696791 | G-A | HET | ||
| SBS | Chr6 | 16289604 | G-A | HET | ||
| SBS | Chr6 | 20065797 | C-T | HET | ||
| SBS | Chr6 | 25712691 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g42770 |
| SBS | Chr6 | 26109660 | G-A | Homo | ||
| SBS | Chr6 | 3717452 | G-A | Homo | ||
| SBS | Chr7 | 10464319 | G-A | Homo | ||
| SBS | Chr8 | 2271016 | T-C | Homo | ||
| SBS | Chr8 | 25590629 | C-A | HET | ||
| SBS | Chr8 | 9563215 | C-T | HET | ||
| SBS | Chr9 | 1351290 | G-T | HET | ||
| SBS | Chr9 | 15572200 | G-A | Homo | ||
| SBS | Chr9 | 15966376 | C-T | Homo | ||
| SBS | Chr9 | 2144189 | C-T | HET | ||
| SBS | Chr9 | 2629915 | C-T | HET | ||
| SBS | Chr9 | 5747252 | C-T | HET | ||
| SBS | Chr9 | 6358052 | G-A | HET | ||
| SBS | Chr9 | 9238426 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g15220 |
| SBS | Chr9 | 9499027 | G-A | Homo |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1193116 | 1193123 | 7 | LOC_Os08g02840 |
| Deletion | Chr8 | 2560912 | 2560913 | 1 | LOC_Os08g04990 |
| Deletion | Chr2 | 5550083 | 5550095 | 12 | |
| Deletion | Chr9 | 5933109 | 5933110 | 1 | |
| Deletion | Chr5 | 8236001 | 8252000 | 15999 | LOC_Os05g14540 |
| Deletion | Chr9 | 9072001 | 9084000 | 11999 | LOC_Os09g15040 |
| Deletion | Chr9 | 9085001 | 9149000 | 63999 | 5 |
| Deletion | Chr9 | 9693256 | 9693257 | 1 | |
| Deletion | Chr4 | 13599912 | 13599913 | 1 | LOC_Os04g23799 |
| Deletion | Chr7 | 19826377 | 19826379 | 2 | |
| Deletion | Chr6 | 25236555 | 25236565 | 10 | |
| Deletion | Chr3 | 25507001 | 25521000 | 13999 | 3 |
| Deletion | Chr5 | 27675781 | 27675785 | 4 | |
| Deletion | Chr3 | 29137034 | 29137038 | 4 | |
| Deletion | Chr3 | 29402503 | 29402505 | 2 | LOC_Os03g51390 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 9515295 | 9515296 | 2 | |
| Insertion | Chr12 | 7961499 | 7961499 | 1 | |
| Insertion | Chr8 | 20947849 | 20947850 | 2 | |
| Insertion | Chr8 | 648057 | 648062 | 6 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 22099731 | 22177457 | 2 |
| Inversion | Chr6 | 25054146 | 26045562 | LOC_Os06g41780 |
| Inversion | Chr6 | 25055310 | 26045632 | LOC_Os06g41780 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 1409116 | Chr1 | 23461414 | 2 |
| Translocation | Chr12 | 5384427 | Chr2 | 16818173 | |
| Translocation | Chr9 | 8943728 | Chr8 | 18611277 | LOC_Os09g14930 |
| Translocation | Chr9 | 9422492 | Chr8 | 18614904 | LOC_Os09g15389 |
| Translocation | Chr11 | 13760012 | Chr6 | 25055305 | 2 |
| Translocation | Chr11 | 13760034 | Chr6 | 25054375 | 2 |
| Translocation | Chr8 | 14223161 | Chr6 | 18930399 | LOC_Os08g23490 |
| Translocation | Chr7 | 17002551 | Chr2 | 17757429 | LOC_Os07g28980 |
| Translocation | Chr11 | 25117154 | Chr1 | 8518603 | |
| Translocation | Chr11 | 25117401 | Chr1 | 8518598 | |
| Translocation | Chr12 | 27012465 | Chr2 | 8113872 |
