Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3527-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3527-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 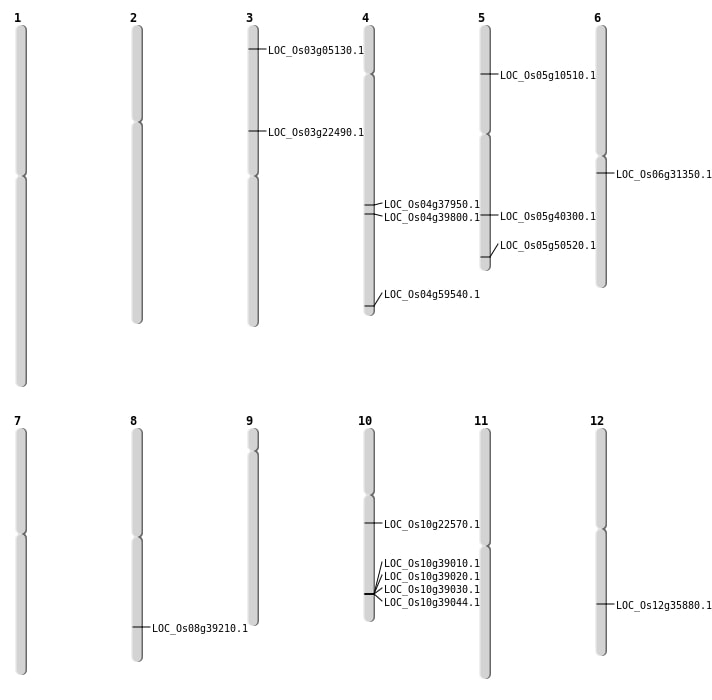
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 3393632 | G-A | Homo | ||
| SBS | Chr1 | 38855779 | G-A | Homo | ||
| SBS | Chr10 | 11358809 | C-T | HET | ||
| SBS | Chr10 | 4827798 | G-A | HET | ||
| SBS | Chr12 | 18237094 | C-T | HET | ||
| SBS | Chr12 | 19375616 | C-T | HET | ||
| SBS | Chr12 | 20036151 | T-C | HET | ||
| SBS | Chr12 | 22716863 | A-T | HET | ||
| SBS | Chr12 | 27524481 | A-T | HET | ||
| SBS | Chr12 | 4955288 | C-T | HET | ||
| SBS | Chr2 | 13578754 | C-T | Homo | ||
| SBS | Chr2 | 21661747 | C-T | HET | ||
| SBS | Chr3 | 12936866 | C-T | HET | ||
| SBS | Chr3 | 23649415 | G-A | HET | ||
| SBS | Chr3 | 2494524 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g05130 |
| SBS | Chr3 | 487107 | C-T | HET | ||
| SBS | Chr4 | 23710287 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g39800 |
| SBS | Chr5 | 28973529 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g50520 |
| SBS | Chr5 | 5610024 | T-C | Homo | ||
| SBS | Chr6 | 12632936 | C-T | HET | ||
| SBS | Chr6 | 30436203 | G-A | Homo | ||
| SBS | Chr7 | 18551286 | G-A | HET | ||
| SBS | Chr7 | 20783800 | G-A | Homo | ||
| SBS | Chr8 | 13777220 | G-A | Homo | ||
| SBS | Chr8 | 24768005 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g39210 |
| SBS | Chr8 | 24768006 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g39210 |
| SBS | Chr8 | 24957989 | A-G | Homo | ||
| SBS | Chr9 | 13577806 | C-A | HET | ||
| SBS | Chr9 | 3455568 | A-C | HET | ||
| SBS | Chr9 | 7161919 | G-C | HET | ||
| SBS | Chr9 | 9017523 | G-A | HET |
Deletions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 909991 | 909994 | 3 | |
| Deletion | Chr5 | 5714544 | 5714550 | 6 | LOC_Os05g10510 |
| Deletion | Chr8 | 9333127 | 9333142 | 15 | |
| Deletion | Chr4 | 15535301 | 15535313 | 12 | |
| Deletion | Chr2 | 17864687 | 17864688 | 1 | |
| Deletion | Chr9 | 18149451 | 18149453 | 2 | |
| Deletion | Chr10 | 20795001 | 20819000 | 23999 | 4 |
| Deletion | Chr5 | 21261795 | 21261796 | 1 | |
| Deletion | Chr2 | 27368794 | 27368800 | 6 | |
| Deletion | Chr5 | 28975276 | 28975284 | 8 | LOC_Os05g50520 |
| Deletion | Chr1 | 32102093 | 32102099 | 6 | |
| Deletion | Chr1 | 41664001 | 41664003 | 2 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 15744837 | 15744837 | 1 | |
| Insertion | Chr12 | 21861348 | 21861351 | 4 | LOC_Os12g35880 |
| Insertion | Chr7 | 13815268 | 13815269 | 2 | |
| Insertion | Chr8 | 6937929 | 6937929 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 5020992 | 5495877 | |
| Inversion | Chr5 | 5471485 | 5495676 | |
| Inversion | Chr3 | 12474128 | 12901531 | |
| Inversion | Chr3 | 12901540 | 12906707 | 2 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 5020537 | Chr4 | 22569086 | 2 |
| Translocation | Chr5 | 5020987 | Chr4 | 22569087 | 2 |
| Translocation | Chr2 | 10394390 | Chr1 | 11768726 | |
| Translocation | Chr10 | 11709917 | Chr2 | 18066714 | LOC_Os10g22570 |
| Translocation | Chr5 | 17627891 | Chr1 | 42980730 | |
| Translocation | Chr6 | 18247541 | Chr4 | 25155880 | LOC_Os06g31350 |
| Translocation | Chr12 | 18408964 | Chr4 | 35415968 | 2 |
| Translocation | Chr10 | 20794575 | Chr5 | 23687789 | 2 |
| Translocation | Chr10 | 20819043 | Chr5 | 23688528 | |
| Translocation | Chr11 | 25810573 | Chr7 | 12810133 | |
| Translocation | Chr6 | 27851559 | Chr3 | 25863274 |
