Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3535-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3535-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 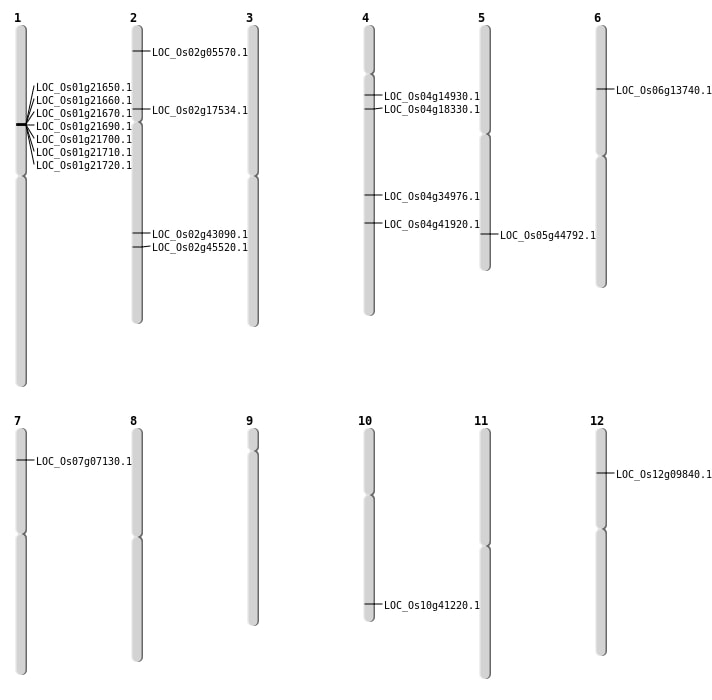
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10918931 | C-T | HET | ||
| SBS | Chr1 | 18830648 | G-A | HET | ||
| SBS | Chr1 | 29806978 | G-A | HET | ||
| SBS | Chr1 | 5431653 | A-T | Homo | ||
| SBS | Chr10 | 15300557 | A-T | HET | ||
| SBS | Chr10 | 888731 | C-T | HET | ||
| SBS | Chr11 | 10751805 | T-C | Homo | ||
| SBS | Chr11 | 11323253 | G-A | HET | ||
| SBS | Chr11 | 11361137 | T-C | Homo | ||
| SBS | Chr11 | 8994086 | C-T | HET | ||
| SBS | Chr12 | 1798446 | G-A | Homo | ||
| SBS | Chr12 | 18725702 | C-G | Homo | ||
| SBS | Chr12 | 20117448 | C-T | Homo | ||
| SBS | Chr12 | 20298999 | A-G | Homo | ||
| SBS | Chr12 | 21803249 | A-G | HET | ||
| SBS | Chr12 | 3114478 | C-T | Homo | ||
| SBS | Chr12 | 5210515 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g09840 |
| SBS | Chr12 | 9030948 | A-C | Homo | ||
| SBS | Chr12 | 9063559 | C-A | Homo | ||
| SBS | Chr2 | 10089453 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g17534 |
| SBS | Chr2 | 19368258 | G-A | HET | ||
| SBS | Chr2 | 335366 | T-C | HET | ||
| SBS | Chr3 | 17229388 | T-A | HET | ||
| SBS | Chr3 | 17229393 | C-A | HET | ||
| SBS | Chr3 | 17273894 | T-G | HET | ||
| SBS | Chr3 | 32311827 | C-T | Homo | ||
| SBS | Chr3 | 4936057 | G-A | HET | ||
| SBS | Chr4 | 10120957 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18330 |
| SBS | Chr4 | 12999371 | G-T | HET | ||
| SBS | Chr4 | 15937683 | C-T | HET | ||
| SBS | Chr4 | 21258675 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g34976 |
| SBS | Chr4 | 24841602 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g41920 |
| SBS | Chr4 | 26293214 | C-T | Homo | ||
| SBS | Chr4 | 8396797 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g14930 |
| SBS | Chr5 | 10750325 | C-A | HET | ||
| SBS | Chr5 | 1623745 | G-A | HET | ||
| SBS | Chr5 | 1944073 | G-C | HET | ||
| SBS | Chr5 | 24010664 | C-T | Homo | ||
| SBS | Chr5 | 26049157 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g44792 |
| SBS | Chr6 | 10687965 | T-C | Homo | ||
| SBS | Chr6 | 27913900 | C-T | HET | ||
| SBS | Chr8 | 18783419 | A-T | HET | ||
| SBS | Chr9 | 13527509 | G-T | Homo | ||
| SBS | Chr9 | 4886230 | A-G | Homo | ||
| SBS | Chr9 | 639284 | C-A | HET | ||
| SBS | Chr9 | 6607532 | C-T | HET |
Deletions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 1873135 | 1873137 | 2 | |
| Deletion | Chr2 | 2706071 | 2706072 | 1 | LOC_Os02g05570 |
| Deletion | Chr9 | 10162636 | 10162637 | 1 | |
| Deletion | Chr1 | 12129001 | 12162000 | 32999 | 3 |
| Deletion | Chr7 | 12166680 | 12166681 | 1 | |
| Deletion | Chr1 | 12168001 | 12188000 | 19999 | 4 |
| Deletion | Chr12 | 14137706 | 14137707 | 1 | |
| Deletion | Chr8 | 15224635 | 15224638 | 3 | |
| Deletion | Chr5 | 17307145 | 17307149 | 4 | |
| Deletion | Chr1 | 21303391 | 21303394 | 3 | |
| Deletion | Chr2 | 27701122 | 27701124 | 2 | LOC_Os02g45520 |
Insertions: 10
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 5934241 | 6421517 | |
| Inversion | Chr6 | 7537988 | 7617590 | 2 |
| Inversion | Chr6 | 7538396 | 7617594 | 2 |
| Inversion | Chr10 | 22138466 | 22138585 | LOC_Os10g41220 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 3524355 | Chr2 | 30308071 | LOC_Os07g07130 |
| Translocation | Chr7 | 3524356 | Chr2 | 18273533 | LOC_Os07g07130 |
| Translocation | Chr7 | 3524370 | Chr2 | 30308540 | LOC_Os07g07130 |
| Translocation | Chr11 | 5818862 | Chr2 | 25950131 | 2 |
| Translocation | Chr9 | 10540260 | Chr5 | 29777448 | |
| Translocation | Chr8 | 11689658 | Chr2 | 13608523 |
