Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3558-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3558-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 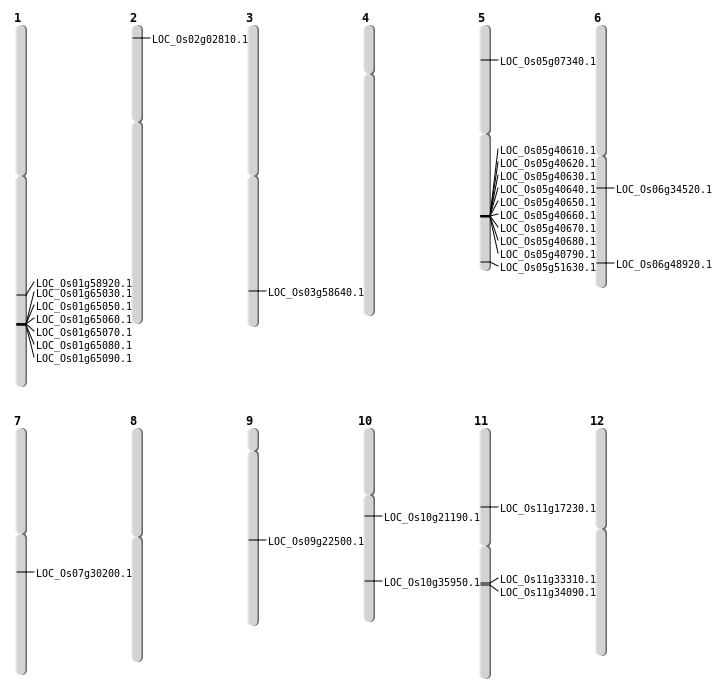
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17483574 | A-T | Homo | ||
| SBS | Chr10 | 10789172 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g21190 |
| SBS | Chr10 | 15597715 | A-G | HET | ||
| SBS | Chr10 | 19214922 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g35950 |
| SBS | Chr11 | 19950299 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g34090 |
| SBS | Chr11 | 19950300 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g34090 |
| SBS | Chr11 | 20768134 | C-T | HET | ||
| SBS | Chr12 | 19809794 | T-C | Homo | ||
| SBS | Chr12 | 2435948 | G-T | HET | ||
| SBS | Chr12 | 4610917 | C-T | HET | ||
| SBS | Chr12 | 9057908 | G-T | HET | ||
| SBS | Chr2 | 1158391 | G-A | HET | ||
| SBS | Chr2 | 1584881 | A-G | HET | ||
| SBS | Chr2 | 21208892 | G-T | HET | ||
| SBS | Chr3 | 32665421 | C-G | Homo | ||
| SBS | Chr3 | 33400815 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g58640 |
| SBS | Chr4 | 1553878 | C-T | Homo | ||
| SBS | Chr4 | 30299053 | T-G | Homo | ||
| SBS | Chr4 | 8278449 | G-A | HET | ||
| SBS | Chr5 | 10815152 | A-G | Homo | ||
| SBS | Chr5 | 29609923 | C-T | Homo | ||
| SBS | Chr5 | 3901110 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g07340 |
| SBS | Chr5 | 3901111 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g07340 |
| SBS | Chr5 | 6573588 | A-G | Homo | ||
| SBS | Chr7 | 16374441 | A-C | Homo | ||
| SBS | Chr7 | 17843911 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30200 |
| SBS | Chr7 | 22722592 | C-T | HET | ||
| SBS | Chr7 | 22722593 | C-G | HET | ||
| SBS | Chr8 | 22564335 | G-T | Homo | ||
| SBS | Chr8 | 22564339 | T-A | Homo | ||
| SBS | Chr9 | 10175955 | A-T | Homo | ||
| SBS | Chr9 | 12145115 | G-A | Homo | ||
| SBS | Chr9 | 13571677 | A-T | HET | STOP_GAINED | LOC_Os09g22500 |
| SBS | Chr9 | 13571678 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g22500 |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1071926 | 1071944 | 18 | LOC_Os02g02810 |
| Deletion | Chr4 | 2797684 | 2797685 | 1 | |
| Deletion | Chr5 | 3902086 | 3902107 | 21 | LOC_Os05g07340 |
| Deletion | Chr5 | 6559322 | 6559328 | 6 | |
| Deletion | Chr5 | 8370184 | 8370188 | 4 | |
| Deletion | Chr11 | 9601880 | 9601883 | 3 | LOC_Os11g17230 |
| Deletion | Chr4 | 10158058 | 10158064 | 6 | |
| Deletion | Chr5 | 10811514 | 10811523 | 9 | |
| Deletion | Chr4 | 15497122 | 15497126 | 4 | |
| Deletion | Chr11 | 16809509 | 16809572 | 63 | |
| Deletion | Chr6 | 20085974 | 20085977 | 3 | LOC_Os06g34520 |
| Deletion | Chr1 | 23074181 | 23074189 | 8 | |
| Deletion | Chr5 | 23818001 | 23856000 | 37999 | 8 |
| Deletion | Chr3 | 25751673 | 25751678 | 5 | |
| Deletion | Chr3 | 26515712 | 26515714 | 2 | |
| Deletion | Chr5 | 27100628 | 27100635 | 7 | |
| Deletion | Chr5 | 27106817 | 27106825 | 8 | |
| Deletion | Chr5 | 29614631 | 29614636 | 5 | LOC_Os05g51630 |
| Deletion | Chr6 | 29619047 | 29619048 | 1 | LOC_Os06g48920 |
| Deletion | Chr4 | 32625102 | 32625109 | 7 | |
| Deletion | Chr1 | 34053453 | 34053497 | 44 | LOC_Os01g58920 |
| Deletion | Chr1 | 37747001 | 37781000 | 33999 | 6 |
Insertions: 7
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 19522241 | 19644478 | |
| Inversion | Chr11 | 19644485 | 19701682 | 2 |
| Inversion | Chr6 | 22483086 | 22521343 | |
| Inversion | Chr5 | 23855091 | 23921060 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 2163974 | Chr1 | 37409969 |
