Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3626-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3626-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 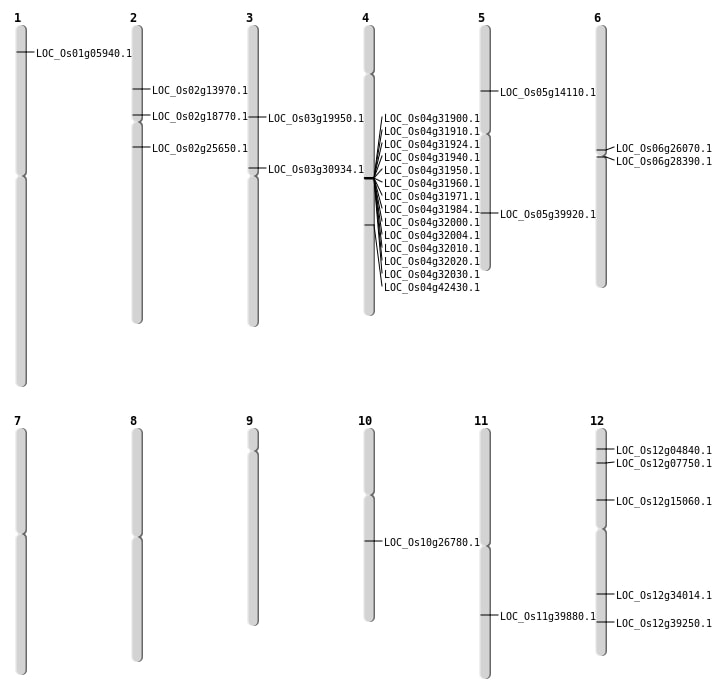
|
| Variant Information |
|---|
Single base substitutions: 55
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23997231 | G-T | HET | ||
| SBS | Chr1 | 26809866 | A-C | HET | ||
| SBS | Chr1 | 27220059 | T-A | HET | ||
| SBS | Chr1 | 2831713 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g05940 |
| SBS | Chr1 | 30427043 | G-A | HET | ||
| SBS | Chr1 | 40371333 | G-T | HET | ||
| SBS | Chr1 | 40493606 | A-G | HET | ||
| SBS | Chr10 | 14086508 | A-C | Homo | ||
| SBS | Chr11 | 14597499 | A-T | HET | ||
| SBS | Chr11 | 18945820 | C-T | HET | ||
| SBS | Chr11 | 21729247 | G-T | HET | ||
| SBS | Chr11 | 8198128 | A-T | Homo | ||
| SBS | Chr12 | 12338060 | G-A | HET | ||
| SBS | Chr12 | 16522270 | C-T | HET | ||
| SBS | Chr12 | 2081416 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g04840 |
| SBS | Chr12 | 20890979 | C-A | HET | ||
| SBS | Chr12 | 3897057 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g07750 |
| SBS | Chr12 | 5260562 | T-A | HET | ||
| SBS | Chr12 | 7978960 | C-A | HET | ||
| SBS | Chr12 | 8594243 | G-A | HET | STOP_GAINED | LOC_Os12g15060 |
| SBS | Chr2 | 12372183 | T-C | HET | ||
| SBS | Chr2 | 12927066 | T-A | HET | ||
| SBS | Chr2 | 15003444 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g25650 |
| SBS | Chr2 | 20468964 | A-T | HET | ||
| SBS | Chr2 | 26421707 | A-G | HET | ||
| SBS | Chr2 | 29654727 | G-T | Homo | ||
| SBS | Chr2 | 8017696 | G-T | HET | ||
| SBS | Chr2 | 8951089 | G-A | HET | ||
| SBS | Chr3 | 10020039 | A-G | HET | ||
| SBS | Chr3 | 35613889 | C-T | Homo | ||
| SBS | Chr3 | 7395705 | C-A | HET | ||
| SBS | Chr4 | 12774809 | G-A | HET | ||
| SBS | Chr4 | 13696780 | C-T | HET | ||
| SBS | Chr4 | 13891326 | C-T | HET | ||
| SBS | Chr4 | 18476934 | C-T | HET | ||
| SBS | Chr4 | 25114831 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g42430 |
| SBS | Chr4 | 28082084 | C-A | HET | ||
| SBS | Chr4 | 28163666 | C-T | HET | ||
| SBS | Chr4 | 4522302 | G-A | HET | ||
| SBS | Chr5 | 6328538 | C-A | HET | ||
| SBS | Chr5 | 7886208 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g14110 |
| SBS | Chr5 | 7886210 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g14110 |
| SBS | Chr6 | 114084 | G-A | HET | ||
| SBS | Chr6 | 15257654 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g26070 |
| SBS | Chr6 | 17335295 | A-C | Homo | ||
| SBS | Chr6 | 7916492 | T-A | Homo | ||
| SBS | Chr7 | 15112563 | G-A | HET | ||
| SBS | Chr7 | 4546561 | A-T | HET | ||
| SBS | Chr8 | 19396395 | C-A | HET | ||
| SBS | Chr8 | 22579056 | A-T | HET | ||
| SBS | Chr8 | 26397992 | T-C | HET | ||
| SBS | Chr9 | 12863094 | A-G | Homo | ||
| SBS | Chr9 | 13459756 | C-T | Homo | ||
| SBS | Chr9 | 1859183 | G-A | Homo | ||
| SBS | Chr9 | 18654006 | G-A | HET |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1910419 | 1910422 | 3 | |
| Deletion | Chr12 | 2082649 | 2082650 | 1 | |
| Deletion | Chr4 | 5023940 | 5023941 | 1 | |
| Deletion | Chr6 | 5718500 | 5718502 | 2 | |
| Deletion | Chr6 | 6671542 | 6671543 | 1 | |
| Deletion | Chr1 | 6798178 | 6798201 | 23 | |
| Deletion | Chr8 | 7385864 | 7385873 | 9 | |
| Deletion | Chr2 | 7392392 | 7392394 | 2 | |
| Deletion | Chr1 | 13925167 | 13925177 | 10 | |
| Deletion | Chr6 | 17377638 | 17377644 | 6 | |
| Deletion | Chr4 | 19103001 | 19193000 | 89999 | 13 |
| Deletion | Chr6 | 19657202 | 19657203 | 1 | |
| Deletion | Chr5 | 23456526 | 23456530 | 4 | LOC_Os05g39920 |
| Deletion | Chr11 | 23773095 | 23773133 | 38 | LOC_Os11g39880 |
| Deletion | Chr7 | 28910080 | 28910087 | 7 | |
| Deletion | Chr2 | 35042698 | 35042702 | 4 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 30195149 | 30195152 | 4 | |
| Insertion | Chr3 | 20263212 | 20263220 | 9 | |
| Insertion | Chr5 | 29945899 | 29945899 | 1 | |
| Insertion | Chr7 | 6425782 | 6425782 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 7625928 | 7926048 | LOC_Os02g13970 |
| Inversion | Chr2 | 7625933 | 7926048 | LOC_Os02g13970 |
| Inversion | Chr1 | 33576537 | 33843301 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2254530 | Chr11 | 22656624 | |
| Translocation | Chr2 | 7625925 | Chr1 | 33843492 | LOC_Os02g13970 |
| Translocation | Chr2 | 7926044 | Chr1 | 33843340 | |
| Translocation | Chr6 | 16151231 | Chr2 | 10939749 | 2 |
| Translocation | Chr4 | 19146895 | Chr3 | 17621955 | LOC_Os03g30934 |
| Translocation | Chr12 | 20529122 | Chr10 | 14013700 | LOC_Os10g26780 |
| Translocation | Chr12 | 20534389 | Chr10 | 14013701 | 2 |
| Translocation | Chr12 | 20944384 | Chr1 | 33576563 | |
| Translocation | Chr12 | 24160557 | Chr3 | 8413098 | LOC_Os12g39250 |
| Translocation | Chr4 | 25757152 | Chr3 | 11224776 | LOC_Os03g19950 |
| Translocation | Chr12 | 26966677 | Chr6 | 4303675 |
