Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3632-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3632-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 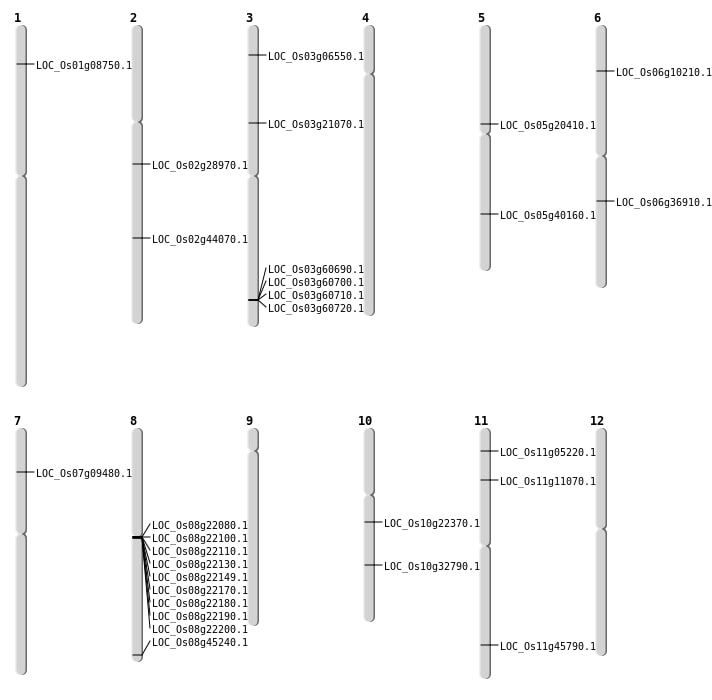
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11103994 | C-T | HET | ||
| SBS | Chr1 | 17374522 | T-A | HET | ||
| SBS | Chr1 | 28986879 | G-A | HET | ||
| SBS | Chr1 | 4383483 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g08750 |
| SBS | Chr10 | 11575072 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g22370 |
| SBS | Chr10 | 17171143 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g32790 |
| SBS | Chr11 | 2314168 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g05220 |
| SBS | Chr11 | 2314169 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g05220 |
| SBS | Chr12 | 2221028 | A-G | HET | ||
| SBS | Chr12 | 26400197 | T-G | Homo | ||
| SBS | Chr2 | 13060267 | A-G | HET | ||
| SBS | Chr2 | 17125692 | G-A | HET | ||
| SBS | Chr2 | 17151748 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g28970 |
| SBS | Chr2 | 21379204 | A-G | HET | ||
| SBS | Chr2 | 24575787 | G-A | Homo | ||
| SBS | Chr2 | 26600783 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g44070 |
| SBS | Chr3 | 13059247 | T-C | HET | ||
| SBS | Chr3 | 15440266 | C-T | HET | ||
| SBS | Chr3 | 21942043 | G-T | HET | ||
| SBS | Chr3 | 29296956 | T-A | HET | ||
| SBS | Chr3 | 6848612 | T-C | HET | ||
| SBS | Chr3 | 6946105 | C-T | HET | ||
| SBS | Chr3 | 9292431 | A-T | HET | ||
| SBS | Chr4 | 1336594 | G-A | HET | ||
| SBS | Chr4 | 18487807 | T-C | HET | ||
| SBS | Chr4 | 23297364 | A-T | Homo | ||
| SBS | Chr4 | 25656932 | G-A | Homo | ||
| SBS | Chr4 | 9335967 | T-C | HET | ||
| SBS | Chr5 | 11973500 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g20410 |
| SBS | Chr6 | 24479789 | G-A | Homo | ||
| SBS | Chr6 | 5230275 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g10210 |
| SBS | Chr6 | 5230276 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g10210 |
| SBS | Chr7 | 14116871 | T-A | HET | ||
| SBS | Chr7 | 16443538 | G-C | Homo | ||
| SBS | Chr7 | 16737687 | T-C | HET | ||
| SBS | Chr7 | 16894448 | C-T | HET | ||
| SBS | Chr7 | 16894466 | C-A | HET | ||
| SBS | Chr7 | 18152219 | C-T | HET | ||
| SBS | Chr7 | 20748453 | C-G | HET | ||
| SBS | Chr7 | 5004942 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g09480 |
| SBS | Chr8 | 11652742 | G-T | HET | ||
| SBS | Chr8 | 17155824 | T-C | HET | ||
| SBS | Chr8 | 18297682 | A-G | HET | ||
| SBS | Chr8 | 21351702 | A-T | HET | ||
| SBS | Chr8 | 25465930 | A-G | HET | ||
| SBS | Chr8 | 3680724 | A-C | Homo | ||
| SBS | Chr9 | 1687017 | T-A | HET | ||
| SBS | Chr9 | 19717688 | T-G | Homo | ||
| SBS | Chr9 | 6364593 | C-A | Homo | ||
| SBS | Chr9 | 8172648 | A-G | HET |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 407020 | 407021 | 1 | |
| Deletion | Chr8 | 2449093 | 2449094 | 1 | |
| Deletion | Chr4 | 2802354 | 2802361 | 7 | |
| Deletion | Chr3 | 3299707 | 3299708 | 1 | LOC_Os03g06550 |
| Deletion | Chr2 | 3991750 | 3991754 | 4 | |
| Deletion | Chr12 | 4391689 | 4391693 | 4 | |
| Deletion | Chr7 | 4925942 | 4925957 | 15 | |
| Deletion | Chr1 | 5108652 | 5108653 | 1 | |
| Deletion | Chr2 | 5187990 | 5187991 | 1 | |
| Deletion | Chr12 | 5394176 | 5394181 | 5 | |
| Deletion | Chr11 | 6121003 | 6121014 | 11 | LOC_Os11g11070 |
| Deletion | Chr10 | 6473780 | 6473781 | 1 | |
| Deletion | Chr3 | 8628023 | 8628037 | 14 | |
| Deletion | Chr7 | 9987441 | 9987445 | 4 | |
| Deletion | Chr8 | 13323001 | 13398000 | 74999 | 9 |
| Deletion | Chr4 | 14673962 | 14673963 | 1 | |
| Deletion | Chr1 | 17955823 | 17955838 | 15 | |
| Deletion | Chr1 | 20347846 | 20347847 | 1 | |
| Deletion | Chr12 | 20352311 | 20352315 | 4 | |
| Deletion | Chr5 | 20494682 | 20494690 | 8 | |
| Deletion | Chr8 | 23356137 | 23356138 | 1 | |
| Deletion | Chr2 | 24651275 | 24651276 | 1 | |
| Deletion | Chr8 | 25954094 | 25954097 | 3 | |
| Deletion | Chr6 | 26432172 | 26432179 | 7 | |
| Deletion | Chr8 | 28407348 | 28407369 | 21 | LOC_Os08g45240 |
| Deletion | Chr2 | 30216881 | 30216882 | 1 | |
| Deletion | Chr2 | 33782173 | 33782179 | 6 | |
| Deletion | Chr3 | 34493001 | 34518000 | 24999 | 4 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 24093373 | 24093373 | 1 | |
| Insertion | Chr12 | 22907764 | 22907765 | 2 | |
| Insertion | Chr8 | 28366463 | 28366463 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 11988134 | 11988433 | LOC_Os03g21070 |
| Inversion | Chr6 | 21751152 | 21751526 | LOC_Os06g36910 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 6265147 | Chr2 | 11770524 | |
| Translocation | Chr6 | 17874850 | Chr5 | 12666964 | |
| Translocation | Chr11 | 27705540 | Chr5 | 23588498 | 2 |
