Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3663-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3663-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 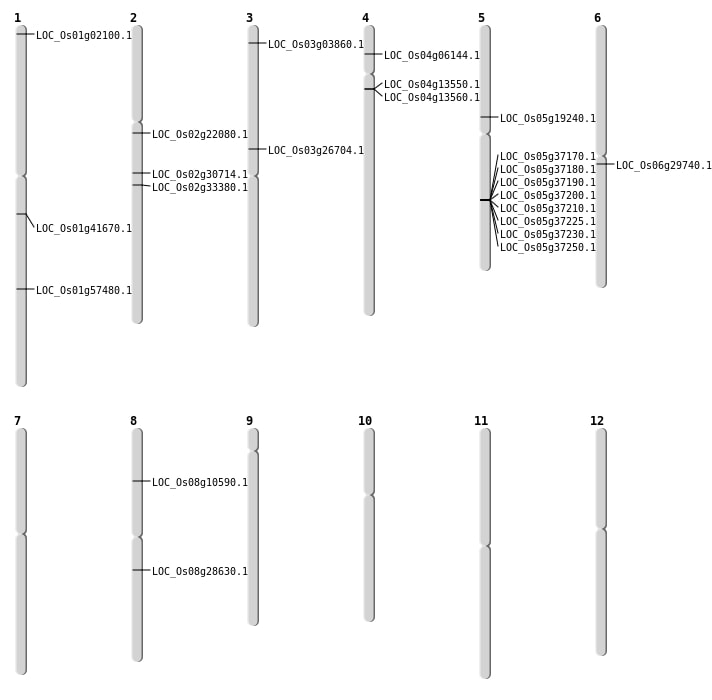
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20461911 | C-T | HET | ||
| SBS | Chr1 | 32860471 | C-T | Homo | ||
| SBS | Chr1 | 36447297 | C-T | HET | ||
| SBS | Chr1 | 391414 | G-A | HET | ||
| SBS | Chr1 | 40258067 | G-A | HET | ||
| SBS | Chr1 | 953574 | C-T | HET | ||
| SBS | Chr10 | 3380259 | G-C | Homo | ||
| SBS | Chr10 | 4275278 | C-T | Homo | ||
| SBS | Chr10 | 4917779 | C-T | HET | ||
| SBS | Chr10 | 6062505 | C-T | HET | ||
| SBS | Chr11 | 15877004 | G-C | HET | ||
| SBS | Chr2 | 18478199 | C-T | HET | ||
| SBS | Chr2 | 19834535 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g33380 |
| SBS | Chr2 | 29298969 | A-G | HET | ||
| SBS | Chr2 | 33919624 | G-A | HET | ||
| SBS | Chr3 | 1739794 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g03860 |
| SBS | Chr3 | 24653671 | C-A | HET | ||
| SBS | Chr3 | 27211480 | C-T | HET | ||
| SBS | Chr3 | 32317252 | A-T | HET | ||
| SBS | Chr4 | 10825907 | G-T | HET | ||
| SBS | Chr4 | 3196971 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g06144 |
| SBS | Chr4 | 4987723 | C-T | HET | ||
| SBS | Chr4 | 748103 | C-T | HET | ||
| SBS | Chr5 | 11174508 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g19240 |
| SBS | Chr5 | 21594896 | T-C | HET | ||
| SBS | Chr6 | 13912810 | C-T | HET | ||
| SBS | Chr6 | 17078529 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g29740 |
| SBS | Chr6 | 7940205 | C-T | HET | ||
| SBS | Chr8 | 14065173 | A-G | HET | ||
| SBS | Chr8 | 5912234 | C-T | HET | ||
| SBS | Chr8 | 6233498 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g10590 |
| SBS | Chr8 | 7725644 | G-A | HET | ||
| SBS | Chr9 | 20791763 | A-G | HET |
Deletions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 4601103 | 4601108 | 5 | |
| Deletion | Chr4 | 7564001 | 7578000 | 13999 | 2 |
| Deletion | Chr2 | 13136192 | 13136195 | 3 | LOC_Os02g22080 |
| Deletion | Chr1 | 14984915 | 14984928 | 13 | |
| Deletion | Chr2 | 15334931 | 15334932 | 1 | |
| Deletion | Chr10 | 15771807 | 15771813 | 6 | |
| Deletion | Chr5 | 21721001 | 21795000 | 73999 | 8 |
| Deletion | Chr1 | 23587384 | 23587391 | 7 | LOC_Os01g41670 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 16256193 | 16256193 | 1 | |
| Insertion | Chr1 | 18289770 | 18289770 | 1 | |
| Insertion | Chr5 | 2697098 | 2697099 | 2 | |
| Insertion | Chr5 | 8423381 | 8423381 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 601335 | 1529953 | LOC_Os01g02100 |
| Inversion | Chr2 | 25882272 | 25882525 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 9838480 | Chr1 | 33213042 | LOC_Os01g57480 |
| Translocation | Chr4 | 9840035 | Chr1 | 33213036 | LOC_Os01g57480 |
| Translocation | Chr7 | 10003290 | Chr2 | 18306591 | LOC_Os02g30714 |
| Translocation | Chr7 | 10003303 | Chr2 | 18305819 | LOC_Os02g30714 |
| Translocation | Chr8 | 17495805 | Chr3 | 15244359 | 2 |
| Translocation | Chr3 | 23252547 | Chr2 | 9805475 | |
| Translocation | Chr11 | 23278362 | Chr5 | 27174266 |
