Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3665-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3665-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 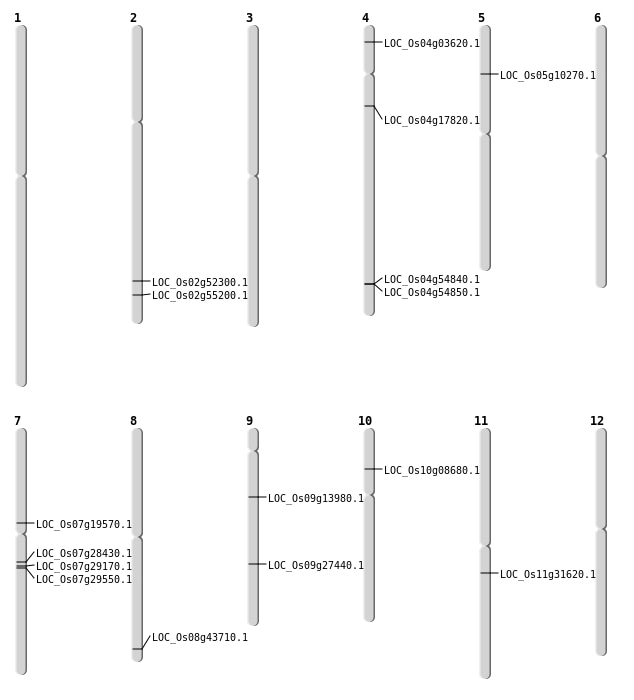
|
| Variant Information |
|---|
Single base substitutions: 58
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11452057 | C-G | HET | ||
| SBS | Chr1 | 15232855 | C-T | HET | ||
| SBS | Chr1 | 1725201 | C-T | HET | ||
| SBS | Chr1 | 32113545 | G-A | HET | ||
| SBS | Chr1 | 34023750 | G-T | HET | ||
| SBS | Chr1 | 36172219 | G-A | HET | ||
| SBS | Chr10 | 13043972 | A-C | HET | ||
| SBS | Chr10 | 14630025 | G-T | HET | ||
| SBS | Chr10 | 14630026 | A-T | HET | ||
| SBS | Chr11 | 778173 | C-T | Homo | ||
| SBS | Chr12 | 14814785 | G-A | HET | ||
| SBS | Chr2 | 12505382 | A-G | HET | ||
| SBS | Chr2 | 12505385 | A-G | HET | ||
| SBS | Chr2 | 24122493 | T-A | Homo | ||
| SBS | Chr2 | 30949005 | C-T | HET | ||
| SBS | Chr2 | 6521546 | G-A | HET | ||
| SBS | Chr3 | 151486 | G-C | Homo | ||
| SBS | Chr3 | 15566908 | G-T | HET | ||
| SBS | Chr3 | 30215260 | T-C | HET | ||
| SBS | Chr3 | 9527691 | T-G | Homo | ||
| SBS | Chr4 | 10029379 | G-A | HET | ||
| SBS | Chr4 | 1582040 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g03620 |
| SBS | Chr4 | 18308708 | G-C | HET | ||
| SBS | Chr4 | 9770136 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g17820 |
| SBS | Chr5 | 12122345 | C-T | HET | ||
| SBS | Chr5 | 16243938 | T-C | HET | ||
| SBS | Chr5 | 2494267 | C-A | Homo | ||
| SBS | Chr5 | 29660178 | G-A | HET | ||
| SBS | Chr5 | 3640426 | A-G | HET | ||
| SBS | Chr5 | 5936897 | A-T | HET | ||
| SBS | Chr6 | 1490666 | C-T | HET | ||
| SBS | Chr6 | 23493382 | G-A | HET | ||
| SBS | Chr6 | 30751176 | T-A | HET | ||
| SBS | Chr6 | 3498837 | G-A | Homo | ||
| SBS | Chr6 | 6959951 | C-T | HET | ||
| SBS | Chr7 | 1097515 | C-A | Homo | ||
| SBS | Chr7 | 11598160 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g19570 |
| SBS | Chr7 | 12647151 | G-T | Homo | ||
| SBS | Chr7 | 14363749 | C-A | HET | ||
| SBS | Chr7 | 16624013 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g28430 |
| SBS | Chr7 | 17080268 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g29170 |
| SBS | Chr7 | 17352227 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g29550 |
| SBS | Chr7 | 22954333 | A-T | HET | ||
| SBS | Chr7 | 2340022 | C-T | HET | ||
| SBS | Chr7 | 6903870 | C-T | HET | ||
| SBS | Chr7 | 7053374 | G-A | HET | ||
| SBS | Chr8 | 18808807 | C-T | Homo | ||
| SBS | Chr8 | 18867882 | C-A | HET | ||
| SBS | Chr8 | 27626866 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g43710 |
| SBS | Chr8 | 9191549 | C-T | Homo | ||
| SBS | Chr9 | 10042623 | G-A | HET | ||
| SBS | Chr9 | 12026143 | G-A | HET | ||
| SBS | Chr9 | 16661422 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g27440 |
| SBS | Chr9 | 3836553 | C-A | HET | ||
| SBS | Chr9 | 4076991 | G-A | HET | ||
| SBS | Chr9 | 5709653 | G-C | HET | ||
| SBS | Chr9 | 8236581 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g13980 |
| SBS | Chr9 | 943430 | A-G | HET |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 111783 | 111792 | 9 | |
| Deletion | Chr2 | 737966 | 737967 | 1 | |
| Deletion | Chr10 | 1400718 | 1400727 | 9 | |
| Deletion | Chr9 | 4138312 | 4138315 | 3 | |
| Deletion | Chr10 | 4711718 | 4711719 | 1 | LOC_Os10g08680 |
| Deletion | Chr5 | 5603717 | 5603718 | 1 | LOC_Os05g10270 |
| Deletion | Chr3 | 5697765 | 5697766 | 1 | |
| Deletion | Chr7 | 10971081 | 10971083 | 2 | |
| Deletion | Chr8 | 14294608 | 14294616 | 8 | |
| Deletion | Chr1 | 14638360 | 14638368 | 8 | |
| Deletion | Chr2 | 15796036 | 15796037 | 1 | |
| Deletion | Chr12 | 16906065 | 16906075 | 10 | |
| Deletion | Chr11 | 23585548 | 23585550 | 2 | |
| Deletion | Chr6 | 23865067 | 23865069 | 2 | |
| Deletion | Chr2 | 25552231 | 25552249 | 18 | |
| Deletion | Chr6 | 27500872 | 27500873 | 1 | |
| Deletion | Chr5 | 28326069 | 28326072 | 3 | |
| Deletion | Chr6 | 28389871 | 28389883 | 12 | |
| Deletion | Chr1 | 29194954 | 29194961 | 7 |
Insertions: 7
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 1476609 | 1580552 | |
| Inversion | Chr11 | 18490100 | 19415029 | LOC_Os11g31620 |
| Inversion | Chr1 | 30860554 | 30992010 | |
| Inversion | Chr1 | 30860563 | 30992022 | |
| Inversion | Chr2 | 32022921 | 32028343 | LOC_Os02g52300 |
| Inversion | Chr2 | 32022929 | 32028366 | LOC_Os02g52300 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 1476624 | Chr3 | 32894166 | |
| Translocation | Chr4 | 1585396 | Chr3 | 32907850 | |
| Translocation | Chr8 | 18521799 | Chr2 | 33811552 | LOC_Os02g55200 |
| Translocation | Chr9 | 19197320 | Chr6 | 5268338 | |
| Translocation | Chr4 | 32615074 | Chr3 | 32894433 | LOC_Os04g54840 |
| Translocation | Chr4 | 32628216 | Chr3 | 32894414 | LOC_Os04g54850 |
