Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3713-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3713-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 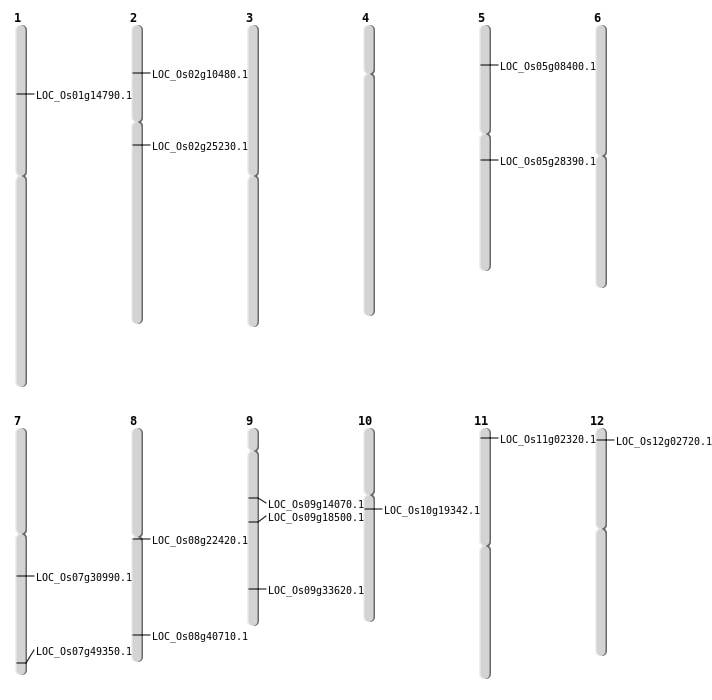
|
| Variant Information |
|---|
Single base substitutions: 47
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17471880 | G-T | HET | ||
| SBS | Chr1 | 24657506 | C-A | HET | ||
| SBS | Chr1 | 31772147 | C-T | HET | ||
| SBS | Chr1 | 8579251 | A-G | HET | ||
| SBS | Chr10 | 15498545 | C-T | Homo | ||
| SBS | Chr11 | 10715677 | A-G | HET | ||
| SBS | Chr11 | 11025354 | A-C | Homo | ||
| SBS | Chr11 | 1121115 | G-A | Homo | ||
| SBS | Chr11 | 12290704 | G-T | Homo | ||
| SBS | Chr11 | 26057909 | T-C | HET | ||
| SBS | Chr11 | 669146 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g02320 |
| SBS | Chr12 | 14478004 | C-T | Homo | ||
| SBS | Chr12 | 652536 | C-A | HET | ||
| SBS | Chr12 | 7436353 | G-T | HET | ||
| SBS | Chr2 | 14682585 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g25230 |
| SBS | Chr2 | 20987953 | G-A | Homo | ||
| SBS | Chr2 | 26164165 | T-C | Homo | ||
| SBS | Chr2 | 5507371 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g10480 |
| SBS | Chr3 | 23164727 | G-T | HET | ||
| SBS | Chr3 | 23552634 | C-T | HET | ||
| SBS | Chr3 | 29227093 | G-A | HET | ||
| SBS | Chr3 | 30909364 | T-C | HET | ||
| SBS | Chr4 | 2063944 | G-T | HET | ||
| SBS | Chr4 | 30602480 | A-G | HET | ||
| SBS | Chr5 | 11273442 | C-T | HET | ||
| SBS | Chr5 | 24724013 | G-C | Homo | ||
| SBS | Chr5 | 28939939 | C-T | HET | ||
| SBS | Chr5 | 4582177 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g08400 |
| SBS | Chr6 | 10857259 | A-G | HET | ||
| SBS | Chr6 | 16937570 | C-T | HET | ||
| SBS | Chr6 | 25314837 | C-A | HET | ||
| SBS | Chr7 | 11512044 | C-T | HET | ||
| SBS | Chr7 | 16271383 | G-A | HET | ||
| SBS | Chr7 | 17623080 | G-T | HET | ||
| SBS | Chr7 | 18355616 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30990 |
| SBS | Chr7 | 2663505 | T-A | HET | ||
| SBS | Chr7 | 26760109 | A-C | HET | ||
| SBS | Chr7 | 28706966 | T-A | Homo | ||
| SBS | Chr7 | 9300138 | C-T | Homo | ||
| SBS | Chr8 | 13526564 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g22420 |
| SBS | Chr8 | 23313878 | G-T | Homo | ||
| SBS | Chr8 | 25768831 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g40710 |
| SBS | Chr9 | 11339492 | G-A | HET | STOP_GAINED | LOC_Os09g18500 |
| SBS | Chr9 | 11765896 | A-T | HET | ||
| SBS | Chr9 | 20008035 | C-A | Homo | ||
| SBS | Chr9 | 21953309 | T-G | HET | ||
| SBS | Chr9 | 8330241 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14070 |
Deletions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 955101 | 955122 | 21 | LOC_Os12g02720 |
| Deletion | Chr2 | 6739780 | 6739781 | 1 | |
| Deletion | Chr10 | 9713150 | 9713153 | 3 | |
| Deletion | Chr1 | 10098199 | 10098202 | 3 | |
| Deletion | Chr5 | 16633005 | 16633024 | 19 | LOC_Os05g28390 |
| Deletion | Chr2 | 16800745 | 16800747 | 2 | |
| Deletion | Chr9 | 19854445 | 19854446 | 1 | LOC_Os09g33620 |
| Deletion | Chr11 | 20303559 | 20303561 | 2 | |
| Deletion | Chr12 | 20935499 | 20935505 | 6 | |
| Deletion | Chr6 | 29867892 | 29867902 | 10 | |
| Deletion | Chr3 | 35725068 | 35725069 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 8272216 | 8272216 | 1 | LOC_Os01g14790 |
| Insertion | Chr11 | 21099236 | 21099238 | 3 | |
| Insertion | Chr8 | 15740446 | 15740447 | 2 |
No Inversion
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2909751 | Chr7 | 29555744 | |
| Translocation | Chr10 | 2915559 | Chr7 | 29557463 | LOC_Os07g49350 |
| Translocation | Chr10 | 9880332 | Chr6 | 4103940 | LOC_Os10g19342 |
| Translocation | Chr6 | 13507630 | Chr1 | 24726876 | |
| Translocation | Chr12 | 27067127 | Chr7 | 4745394 |
