Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3723-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3723-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 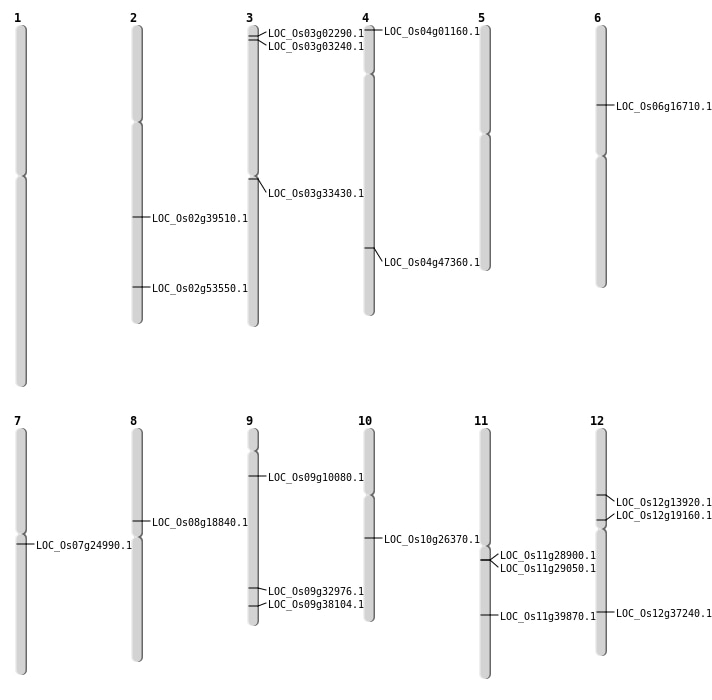
|
| Variant Information |
|---|
Single base substitutions: 51
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13237725 | T-A | HET | ||
| SBS | Chr1 | 13237726 | T-A | HET | ||
| SBS | Chr1 | 16375064 | C-T | HET | ||
| SBS | Chr1 | 16767209 | C-T | HET | ||
| SBS | Chr1 | 17801500 | T-G | HET | ||
| SBS | Chr1 | 17833819 | G-T | Homo | ||
| SBS | Chr1 | 20335370 | C-T | Homo | ||
| SBS | Chr1 | 39903016 | A-G | HET | ||
| SBS | Chr10 | 10969504 | G-A | HET | ||
| SBS | Chr10 | 11129572 | T-C | HET | ||
| SBS | Chr10 | 13376507 | A-G | HET | ||
| SBS | Chr11 | 10491332 | A-T | Homo | ||
| SBS | Chr11 | 11477660 | G-A | Homo | ||
| SBS | Chr11 | 23768054 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g39870 |
| SBS | Chr11 | 9280386 | A-T | Homo | ||
| SBS | Chr12 | 11124013 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g19160 |
| SBS | Chr12 | 15000199 | T-A | HET | ||
| SBS | Chr12 | 22844401 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g37240 |
| SBS | Chr2 | 22819290 | T-A | HET | ||
| SBS | Chr2 | 30903765 | G-A | HET | ||
| SBS | Chr2 | 32764606 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g53550 |
| SBS | Chr2 | 35559600 | T-G | HET | ||
| SBS | Chr2 | 35858559 | C-T | HET | ||
| SBS | Chr2 | 5619965 | G-A | HET | ||
| SBS | Chr2 | 85012 | C-T | Homo | ||
| SBS | Chr2 | 9315468 | C-T | HET | ||
| SBS | Chr3 | 19104982 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g33430 |
| SBS | Chr3 | 22551117 | G-A | HET | ||
| SBS | Chr3 | 23059462 | G-T | HET | ||
| SBS | Chr3 | 23273744 | C-T | HET | ||
| SBS | Chr3 | 23409030 | A-G | HET | ||
| SBS | Chr3 | 31822113 | T-A | HET | ||
| SBS | Chr3 | 36087345 | G-A | HET | ||
| SBS | Chr3 | 7583379 | C-A | Homo | ||
| SBS | Chr4 | 28511350 | A-C | HET | ||
| SBS | Chr4 | 9488160 | A-G | HET | ||
| SBS | Chr5 | 10488803 | C-T | HET | ||
| SBS | Chr5 | 16747631 | A-C | Homo | ||
| SBS | Chr5 | 16747633 | A-G | Homo | ||
| SBS | Chr5 | 3405722 | G-A | HET | ||
| SBS | Chr5 | 4818196 | A-T | HET | ||
| SBS | Chr5 | 9408122 | T-G | HET | ||
| SBS | Chr6 | 10458035 | A-G | HET | ||
| SBS | Chr6 | 16196322 | A-T | Homo | ||
| SBS | Chr6 | 4401149 | C-T | Homo | ||
| SBS | Chr6 | 4401150 | C-T | Homo | ||
| SBS | Chr7 | 24088106 | G-A | Homo | ||
| SBS | Chr9 | 16879348 | T-C | HET | ||
| SBS | Chr9 | 17230124 | C-T | Homo | ||
| SBS | Chr9 | 5495450 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g10080 |
| SBS | Chr9 | 7486809 | G-C | HET |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 728687 | 728688 | 1 | |
| Deletion | Chr3 | 791254 | 791258 | 4 | LOC_Os03g02290 |
| Deletion | Chr11 | 1939908 | 1939909 | 1 | |
| Deletion | Chr7 | 3386242 | 3386253 | 11 | |
| Deletion | Chr9 | 8848409 | 8848410 | 1 | |
| Deletion | Chr9 | 9553098 | 9553108 | 10 | |
| Deletion | Chr9 | 10136507 | 10136508 | 1 | |
| Deletion | Chr3 | 10913274 | 10913275 | 1 | |
| Deletion | Chr8 | 11245192 | 11246187 | 995 | LOC_Os08g18840 |
| Deletion | Chr7 | 12241232 | 12241863 | 631 | |
| Deletion | Chr10 | 13687951 | 13687952 | 1 | LOC_Os10g26370 |
| Deletion | Chr7 | 14228715 | 14228718 | 3 | LOC_Os07g24990 |
| Deletion | Chr7 | 15457941 | 15457943 | 2 | |
| Deletion | Chr11 | 16825732 | 16825735 | 3 | LOC_Os11g29050 |
| Deletion | Chr2 | 23845822 | 23845831 | 9 | LOC_Os02g39510 |
| Deletion | Chr4 | 28109996 | 28109998 | 2 | LOC_Os04g47360 |
| Deletion | Chr3 | 34498436 | 34498437 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 27283855 | 27283863 | 9 | |
| Insertion | Chr4 | 27510253 | 27510253 | 1 | |
| Insertion | Chr9 | 15428300 | 15428301 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 19669841 | 19684281 | LOC_Os09g32976 |
Translocations: 14
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 31 | Chr1 | 1116 | |
| Translocation | Chr8 | 1207 | Chr2 | 151 | |
| Translocation | Chr4 | 122476 | Chr2 | 21182843 | LOC_Os04g01160 |
| Translocation | Chr11 | 2499158 | Chr8 | 19698850 | |
| Translocation | Chr9 | 3258599 | Chr2 | 20742908 | |
| Translocation | Chr4 | 3954787 | Chr3 | 1385060 | LOC_Os03g03240 |
| Translocation | Chr10 | 4400271 | Chr6 | 9639811 | LOC_Os06g16710 |
| Translocation | Chr12 | 7904389 | Chr4 | 18473924 | LOC_Os12g13920 |
| Translocation | Chr12 | 7904396 | Chr4 | 18596360 | LOC_Os12g13920 |
| Translocation | Chr9 | 15007536 | Chr1 | 12925742 | |
| Translocation | Chr11 | 16725357 | Chr10 | 7882591 | LOC_Os11g28900 |
| Translocation | Chr9 | 21954634 | Chr6 | 6426492 | LOC_Os09g38104 |
| Translocation | Chr4 | 33522784 | Chr2 | 21183331 | |
| Translocation | Chr4 | 33522819 | Chr2 | 21182843 |
