Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3729-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3729-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 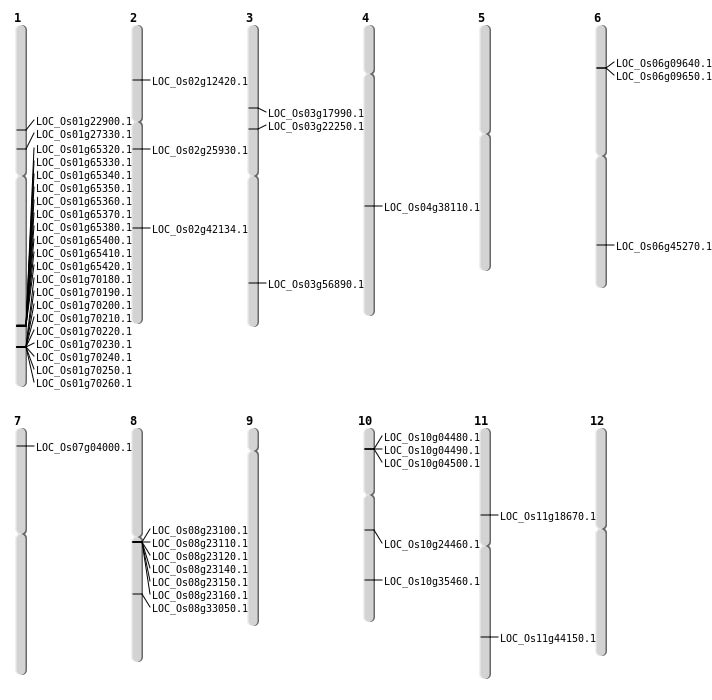
|
| Variant Information |
|---|
Single base substitutions: 47
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14850967 | A-G | HET | ||
| SBS | Chr1 | 15253627 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g27330 |
| SBS | Chr10 | 11521499 | C-T | HET | ||
| SBS | Chr10 | 12553367 | C-A | HET | STOP_GAINED | LOC_Os10g24460 |
| SBS | Chr11 | 11462474 | C-T | HET | ||
| SBS | Chr11 | 17520336 | T-C | Homo | ||
| SBS | Chr11 | 25306498 | C-T | HET | ||
| SBS | Chr11 | 26653208 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g44150 |
| SBS | Chr11 | 27683590 | T-A | HET | ||
| SBS | Chr11 | 4474549 | C-A | HET | ||
| SBS | Chr12 | 10847767 | C-T | HET | ||
| SBS | Chr12 | 1898014 | C-T | Homo | ||
| SBS | Chr12 | 2391088 | T-A | Homo | ||
| SBS | Chr2 | 10551858 | C-T | HET | ||
| SBS | Chr2 | 20261804 | G-A | HET | ||
| SBS | Chr2 | 31811175 | C-T | HET | ||
| SBS | Chr2 | 32571835 | C-T | HET | ||
| SBS | Chr2 | 6475516 | T-G | HET | ||
| SBS | Chr2 | 6475771 | G-T | HET | ||
| SBS | Chr2 | 6477315 | A-T | HET | STOP_GAINED | LOC_Os02g12420 |
| SBS | Chr3 | 10016861 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g17990 |
| SBS | Chr3 | 19468537 | C-T | HET | ||
| SBS | Chr3 | 28216393 | C-T | HET | ||
| SBS | Chr3 | 29516528 | C-A | HET | ||
| SBS | Chr4 | 10934008 | G-A | HET | ||
| SBS | Chr4 | 12623543 | A-G | HET | ||
| SBS | Chr4 | 15061731 | T-A | HET | ||
| SBS | Chr4 | 2163939 | G-T | HET | ||
| SBS | Chr4 | 22681459 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g38110 |
| SBS | Chr4 | 2890390 | C-T | HET | ||
| SBS | Chr4 | 6218474 | C-T | HET | ||
| SBS | Chr5 | 14180433 | G-T | Homo | ||
| SBS | Chr6 | 1709636 | T-C | HET | ||
| SBS | Chr6 | 21266575 | G-A | HET | ||
| SBS | Chr6 | 2617627 | C-T | HET | ||
| SBS | Chr6 | 27357754 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g45270 |
| SBS | Chr7 | 10314366 | C-T | Homo | ||
| SBS | Chr7 | 466524 | C-T | HET | ||
| SBS | Chr7 | 4875765 | C-T | HET | ||
| SBS | Chr7 | 8091973 | C-T | HET | ||
| SBS | Chr7 | 8752087 | A-G | HET | ||
| SBS | Chr8 | 1358311 | C-T | HET | ||
| SBS | Chr8 | 20214784 | G-A | HET | ||
| SBS | Chr8 | 6524944 | T-C | Homo | ||
| SBS | Chr9 | 10913480 | T-C | Homo | ||
| SBS | Chr9 | 21025382 | C-T | Homo | ||
| SBS | Chr9 | 21855385 | A-G | Homo |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 2115001 | 2137000 | 21999 | 3 |
| Deletion | Chr2 | 4065306 | 4065313 | 7 | |
| Deletion | Chr6 | 4905001 | 4915000 | 9999 | 2 |
| Deletion | Chr9 | 7846188 | 7846200 | 12 | |
| Deletion | Chr1 | 10477602 | 10477604 | 2 | |
| Deletion | Chr11 | 10546001 | 10562000 | 15999 | LOC_Os11g18670 |
| Deletion | Chr10 | 12475764 | 12475770 | 6 | |
| Deletion | Chr1 | 12870884 | 12870885 | 1 | LOC_Os01g22900 |
| Deletion | Chr6 | 12983979 | 12983981 | 2 | |
| Deletion | Chr8 | 13910001 | 13972000 | 61999 | 7 |
| Deletion | Chr1 | 16327614 | 16327621 | 7 | |
| Deletion | Chr8 | 20530861 | 20530871 | 10 | LOC_Os08g33050 |
| Deletion | Chr1 | 23210947 | 23210949 | 2 | LOC_Os01g40990 |
| Deletion | Chr3 | 25045492 | 25045507 | 15 | |
| Deletion | Chr4 | 26984149 | 26984156 | 7 | |
| Deletion | Chr1 | 37892001 | 37971000 | 78999 | 10 |
| Deletion | Chr1 | 40290190 | 40290191 | 1 | |
| Deletion | Chr1 | 40592403 | 40592404 | 1 | |
| Deletion | Chr1 | 40617001 | 40683000 | 65999 | 9 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr5 | 10133374 | 10133375 | 2 | |
| Insertion | Chr6 | 1309350 | 1309351 | 2 | |
| Insertion | Chr7 | 11884 | 11885 | 2 | |
| Insertion | Chr7 | 20126629 | 20126630 | 2 |
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 12892528 | 12892655 | |
| Inversion | Chr5 | 14494984 | 14495368 | |
| Inversion | Chr2 | 15217180 | 15217543 | LOC_Os02g25930 |
| Inversion | Chr9 | 15820922 | 15821104 | |
| Inversion | Chr5 | 17875068 | 17875280 | |
| Inversion | Chr10 | 18978051 | 18978384 | LOC_Os10g35460 |
| Inversion | Chr4 | 20356108 | 20356493 | |
| Inversion | Chr3 | 30563866 | 30564013 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 1418022 | Chr3 | 32400347 | LOC_Os03g56890 |
| Translocation | Chr5 | 5022236 | Chr2 | 25341897 | LOC_Os02g42134 |
| Translocation | Chr9 | 13581691 | Chr3 | 12759727 | LOC_Os03g22250 |
| Translocation | Chr12 | 17684705 | Chr7 | 1689662 | LOC_Os07g04000 |
